Download some paired-end sequencing data in a FASTQ format using an accession run number SRR11846824 that has two reads per spot and has no unaligned reads. Downloading the data may take a few minutes.
tbl=1×2 table
Reads_1 Reads_2
_____________________ _____________________
SRR11846824 "SRR11846824_1.fastq" "SRR11846824_2.fastq"
By default, the function uses the SplitType="SplitThree" option and downloads only biological reads. Specifically, the function splits spots into reads. For spots having two reads, the function produces *_1.fastq and *_2.fastq, represented by the Reads_1 and Reads_2 columns. If there are any unaligned reads, the function saves unaligned reads in a *.fastq file, which would be represented by the Reads column. Because there are no unaligned reads within this accession, the function did not produce a *.fastq file, and the output table has no Reads column. For details, see SplitType.
You can also specify other download options using SRAFasterqDumpOptions. For instance, use FastaOutput=true to get the FASTA-formatted file.
Alternatively, you can specify the options as name-value arguments instead of using the options object.
tbl2 = srafasterqdump("SRR11846824",FastaOutput=true);
You can also download the data in a SAM format using srasamdump.
samFile =
"SRR11846824.sam"
Specify the download options using an SRASAMDumpOptions object. For instance, specify the output file name and compress the output file using bzip2.
samdumpopt =
SRASAMDumpOptions with properties:
Default properties:
ExtraCommand: ""
FastaOutput: 0
FastqOutput: 0
GZip: 0
HideIdentical: 0
IncludeAll: 0
MinMapQuality: 0
OutputPrimary: 0
OutputUnaligned: 0
Version: "3.0.6"
Modified properties:
OutputFileName: "SRR11846824.sam.bz2"
BZip2: 1
bzFile =
"SRR11846824.sam.bz2"
After downloading the SAM file, you can use it for downstream analyses. For instance, you can use bowtie2 to map the reads to the reference sequence.
First, download the C. elegans reference sequence.
Save Chromosome 3 reference data in a FASTA file.
Build a set of index files using bowtie2build. The status value of 0 means that the build was successful.
Align read data to the reference. This may take a few minutes.
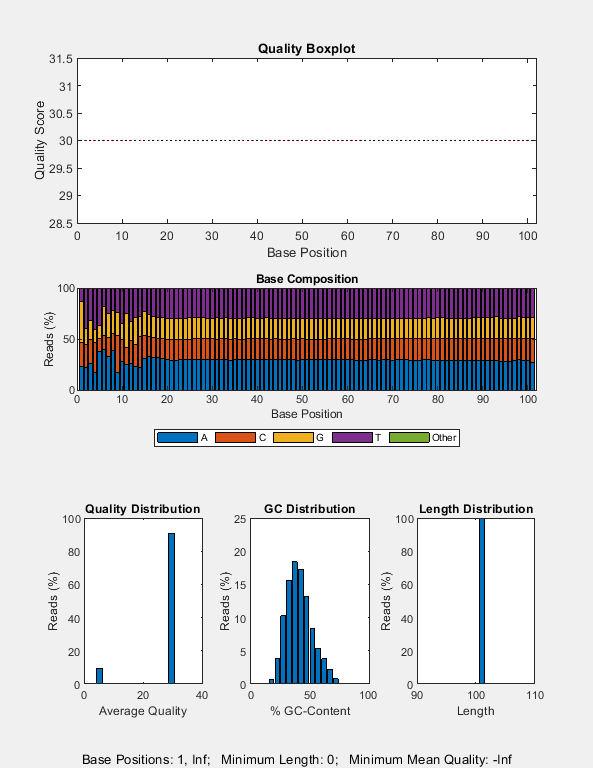
Create a quality control plot for the SAM file. Note that, for this particular experiment, most of the reads happen to have the same quality score of 30.
Convert the SAM file to a BAM file. Suppress two informational warnings that are issued while creating a BioMap object.
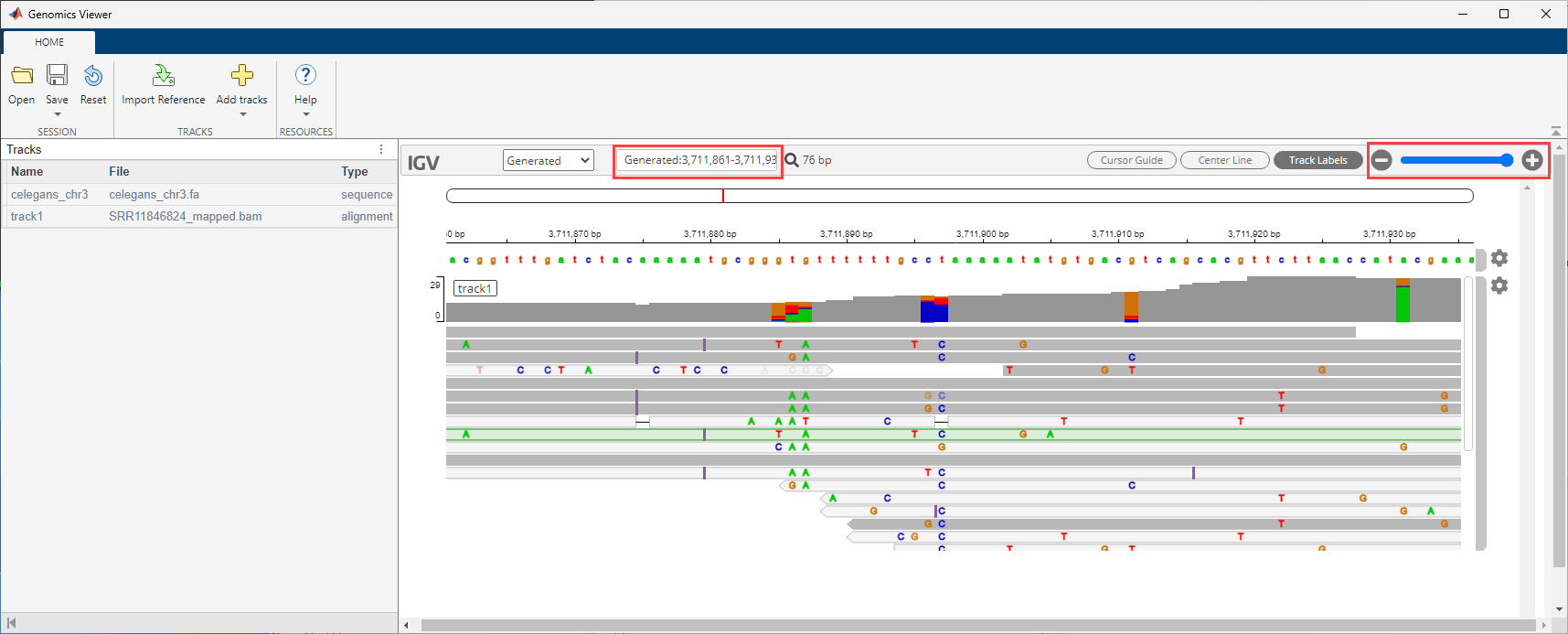
Visualize the alignment data in the Genomics Viewer app. The corresponding cytoband file is provided with the toolbox.
Use the zoom slider to zoom in and see the features. Or you can enter the following in the search text box: Generated:3,711,861-3,711,940.
You may delete the downloaded files, such as the reference sequence file.
Close the app.