使用物理信息神经网络求解 ODE
此示例说明如何训练物理信息神经网络 (PINN) 来预测常微分方程 (ODE) 的解。
并非所有微分方程都有闭式解。为了找到这些类型的方程的近似解,有许多传统的数值算法可供使用。物理信息神经网络 (PINN) [1] 是一种将物理定律融入其结构体和训练过程的神经网络。您可以训练一个神经网络,输出定义物理系统的 ODE 的解。这种方法具有诸多优势,包括以封闭解析形式提供可微分的近似解 [2]。
此示例说明如何完成以下操作:
生成 范围内的训练数据。
定义一个神经网络,该网络以 为输入,并返回在 处计算的 ODE 的近似解作为输出。
使用自定义损失函数训练网络。
将网络预测结果与解析解进行比较。
ODE 和损失函数
在此示例中,您将求解 ODE
初始条件为 。此 ODE 的解析解为
定义一个自定义损失函数,该函数对不满足 ODE 和初始条件的偏差进行罚分。在此示例中,损失函数是 ODE 损失和初始条件损失的加权和:
是网络参数, 是常量系数, 是网络预测的解, 是使用自动微分计算的预测解的导数。 项是 ODE 损失,它量化了预测解与满足 ODE 定义的偏差程度。 项是初始条件损失,它量化了预测解与满足初始条件的偏差程度。
生成输入数据并定义网络
在 范围内生成 10,000 个训练数据点。
x = linspace(0,2,10000)';
定义用于求解 ODE 的网络。由于输入是实数 ,因此指定输入大小为 1。
inputSize = 1;
layers = [
featureInputLayer(inputSize,Normalization="none")
fullyConnectedLayer(10)
sigmoidLayer
fullyConnectedLayer(1)
sigmoidLayer];基于层数组创建一个 dlnetwork 对象。
net = dlnetwork(layers)
net =
dlnetwork with properties:
Layers: [5×1 nnet.cnn.layer.Layer]
Connections: [4×2 table]
Learnables: [4×3 table]
State: [0×3 table]
InputNames: {'input'}
OutputNames: {'layer_2'}
Initialized: 1
View summary with summary.
定义模型损失函数
创建在示例的模型损失函数部分列出的函数 modelLoss,该函数以神经网络、小批量输入数据和与初始条件损失关联的系数为输入。此函数返回损失以及该损失关于神经网络中可学习参数的梯度。
指定训练选项
使用 100 的小批量大小进行 15 轮训练。
numEpochs = 15; miniBatchSize = 100;
指定 SGDM 优化的选项。指定学习率为 0.5、学习率下降因子为 0.5、学习率下降周期为 5、动量为 0.9。
initialLearnRate = 0.5; learnRateDropFactor = 0.5; learnRateDropPeriod = 5; momentum = 0.9;
指定损失函数中初始条件项的系数为 7。此系数指定了初始条件对损失的相对贡献。调整此参数可以帮助训练更快地收敛。
icCoeff = 7;
训练模型
要在训练期间使用小批量数据,请执行以下操作:
基于训练数据创建一个
arrayDatastore对象。创建一个以
arrayDatastore对象为输入的minibatchqueue对象,指定小批量大小以丢弃部分小批量,并且训练数据的格式为"BC"(批量、通道)。
默认情况下,minibatchqueue 对象将数据转换为基础类型为 single 的 dlarray 对象。如果 GPU 可用,该对象还会将每个输出都转换为一个 gpuArray。使用 GPU 需要 Parallel Computing Toolbox™ 和支持的 GPU 设备。有关受支持设备的信息,请参阅GPU 计算要求 (Parallel Computing Toolbox)。
ads = arrayDatastore(x,IterationDimension=1); mbq = minibatchqueue(ads, ... MiniBatchSize=miniBatchSize, ... PartialMiniBatch="discard", ... MiniBatchFormat="BC");
初始化 SGDM 求解器的速度参数。
velocity = [];
计算训练进度监视器的总迭代次数。
numObservationsTrain = numel(x); numIterationsPerEpoch = floor(numObservationsTrain / miniBatchSize); numIterations = numEpochs * numIterationsPerEpoch;
初始化 TrainingProgressMonitor 对象。由于计时器在您创建监视器对象时启动,请确保您创建的对象靠近训练循环。
monitor = trainingProgressMonitor( ... Metrics="LogLoss", ... Info=["Epoch" "LearnRate"], ... XLabel="Iteration");
使用自定义训练循环来训练网络。对于每轮训练,对数据进行乱序处理,并遍历小批量数据。对于每个小批量:
使用
dlfeval和modelLoss函数计算模型损失和梯度。使用
sgdmupdate函数更新网络参数。显示训练进度。
如果
Stop属性为true,则停止训练。当您点击“停止”按钮时,TrainingProgressMonitor对象的Stop属性值会更改为 true。
每 learnRateDropPeriod 轮将学习率乘以 learnRateDropFactor。
epoch = 0; iteration = 0; learnRate = initialLearnRate; start = tic; % Loop over epochs. while epoch < numEpochs && ~monitor.Stop epoch = epoch + 1; % Shuffle data. mbq.shuffle % Loop over mini-batches. while hasdata(mbq) && ~monitor.Stop iteration = iteration + 1; % Read mini-batch of data. X = next(mbq); % Evaluate the model gradients and loss using dlfeval and the modelLoss function. [loss,gradients] = dlfeval(@modelLoss, net, X, icCoeff); % Update network parameters using the SGDM optimizer. [net,velocity] = sgdmupdate(net,gradients,velocity,learnRate,momentum); % Update the training progress monitor. recordMetrics(monitor,iteration,LogLoss=log(loss)); updateInfo(monitor,Epoch=epoch,LearnRate=learnRate); monitor.Progress = 100 * iteration/numIterations; end % Reduce the learning rate. if mod(epoch,learnRateDropPeriod)==0 learnRate = learnRate*learnRateDropFactor; end end
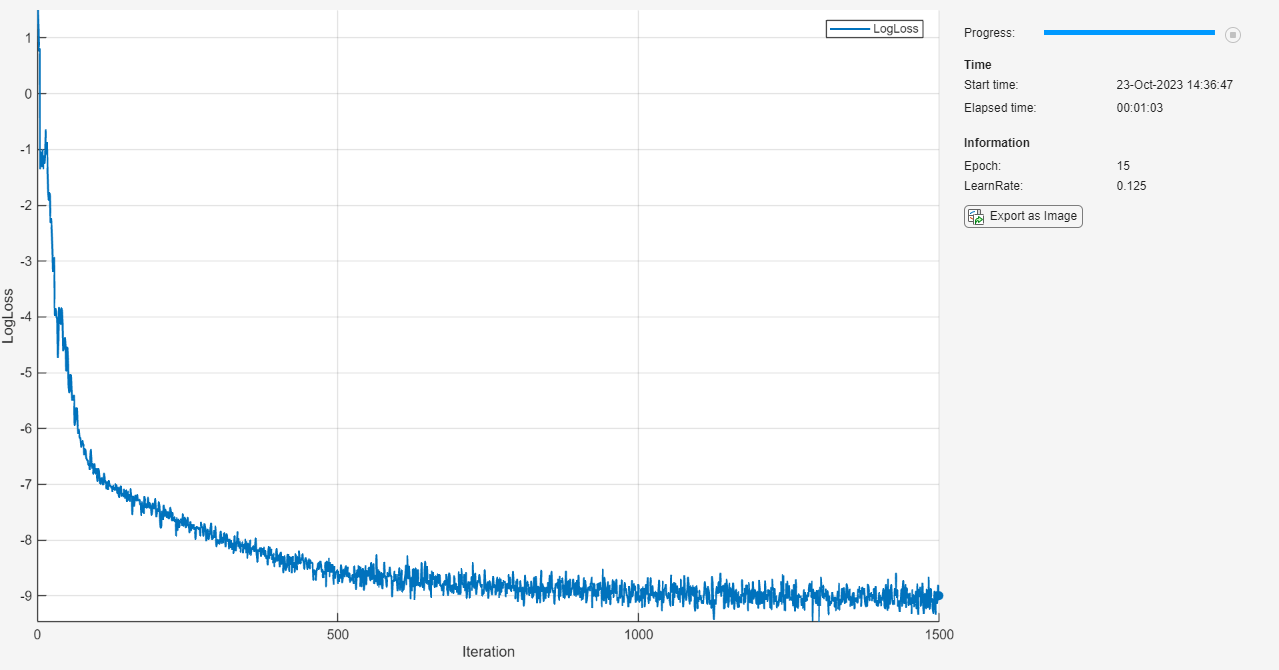
测试模型
通过将网络预测结果与解析解进行比较来测试网络的准确度。
生成 范围内的测试数据,以查看网络是否能够在训练范围 之外进行外插。
xTest = linspace(0,4,1000)';
使用 minibatchpredict 函数进行预测。默认情况下,minibatchpredict 函数使用 GPU(如果有)。使用 GPU 需要 Parallel Computing Toolbox™ 许可证和受支持的 GPU 设备。有关受支持设备的信息,请参阅GPU 计算要求 (Parallel Computing Toolbox)。否则,该函数使用 CPU。要指定执行环境,请使用 ExecutionEnvironment 选项。
yModel = minibatchpredict(net,xTest);
计算解析解。
yAnalytic = exp(-xTest.^2);
通过以对数刻度绘制解析解和模型预测来对它们进行比较。
figure; plot(xTest,yAnalytic,"-") hold on plot(xTest,yModel,"--") legend("Analytic","Model") xlabel("x") ylabel("y (log scale)") set(gca,YScale="log")

模型在训练范围 内准确地逼近解析解,并在 范围内进行外插,但准确度较低。
计算训练范围 内的均方相对误差。
yModelTrain = yModel(1:500); yAnalyticTrain = yAnalytic(1:500); errorTrain = mean(((yModelTrain-yAnalyticTrain)./yAnalyticTrain).^2)
errorTrain = single
0.0029
计算外插范围 内的均方相对误差。
yModelExtra = yModel(501:1000); yAnalyticExtra = yAnalytic(501:1000); errorExtra = mean(((yModelExtra-yAnalyticExtra)./yAnalyticExtra).^2)
errorExtra = single
6.9634e+05
请注意,外插范围内的均方相对误差高于训练范围内的均方相对误差。
模型损失函数
modelLoss 函数以 dlnetwork 对象 net、小批量输入数据 X 和与初始条件损失关联的系数 icCoeff 为输入。此函数返回损失关于 net 中可学习参数的梯度以及相应损失。损失定义为 ODE 损失和初始条件损失的加权和。计算此损失需要二阶导数。要能够进行二阶自动微分,请使用函数 dlgradient 并将 EnableHigherDerivatives 名称值参量设置为 true。
function [loss,gradients] = modelLoss(net, X, icCoeff) y = forward(net,X); % Evaluate the gradient of y with respect to x. % Since another derivative will be taken, set EnableHigherDerivatives to true. dy = dlgradient(sum(y,"all"),X,EnableHigherDerivatives=true); % Define ODE loss. eq = dy + 2*y.*X; % Define initial condition loss. ic = forward(net,dlarray(0,"CB")) - 1; % Specify the loss as a weighted sum of the ODE loss and the initial condition loss. loss = mean(eq.^2,"all") + icCoeff * ic.^2; % Evaluate model gradients. gradients = dlgradient(loss, net.Learnables); end
参考资料
Maziar Raissi, Paris Perdikaris, and George Em Karniadakis, Physics Informed Deep Learning (Part I):Data-driven Solutions of Nonlinear Partial Differential Equations https://arxiv.org/abs/1711.10561
Lagaris, I. E., A. Likas, and D. I. Fotiadis.“Artificial Neural Networks for Solving Ordinary and Partial Differential Equations.”IEEE Transactions on Neural Networks 9, no. 5 (September 1998):987–1000. https://doi.org/10.1109/72.712178.
另请参阅
dlarray | dlfeval | dlgradient