plot
Visualize prior and posterior densities of Bayesian linear regression model parameters
Syntax
Description
plot( or
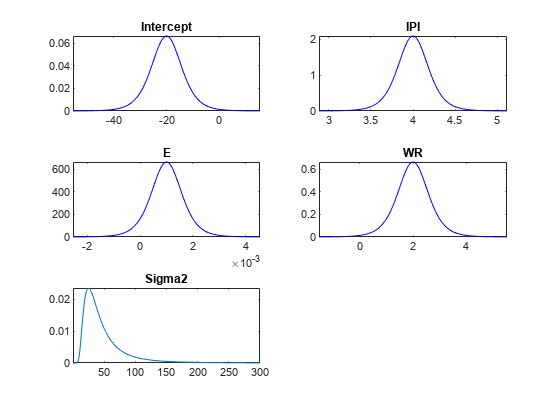
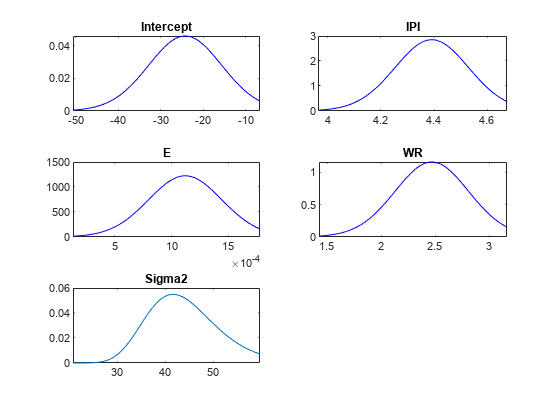
PosteriorMdl)plot( plots the posterior or
prior distributions of the parameters in the Bayesian linear regression
model
PriorMdl)PosteriorMdl or PriorMdl, respectively.
plot adds subplots for each parameter to one figure and
overwrites the same figure when you call plot multiple
times.
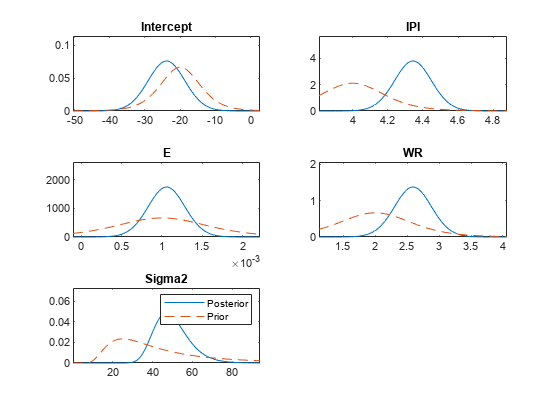
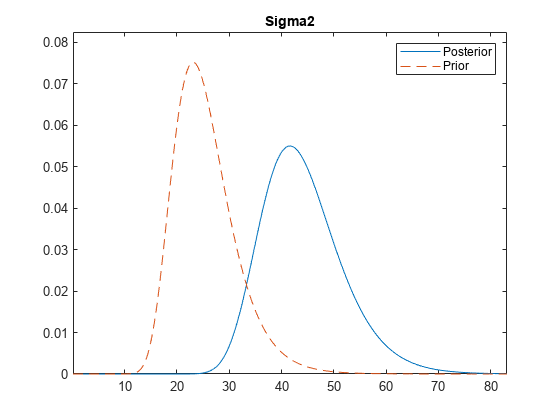
plot(
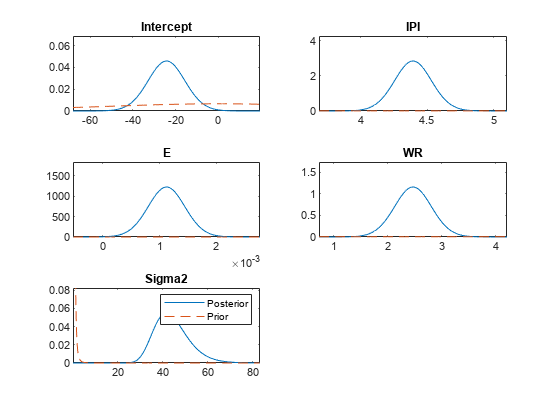
plots the posterior and prior distributions in the same subplot.
PosteriorMdl,PriorMdl)plot uses solid blue lines for posterior
densities and dashed red lines for prior densities.
plot(___,
uses any of the input argument combinations in the previous syntaxes and
additional options specified by one or more name-value pair arguments. For
example, you can evaluate the posterior or prior density by supplying values of
β and σ2,
or choose which parameter distributions to include in the figure.Name,Value)
pointsUsed = plot(___)plot
uses to evaluate the densities in the subplots.
[
also returns the values of the evaluated densities.pointsUsed,posteriorDensity,priorDensity]
= plot(___)
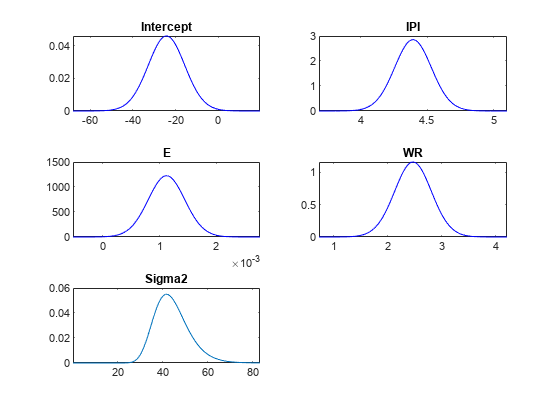
If you specify one model, then plot returns the
density values in PosteriorDensity. Otherwise,
plot returns the posterior density values in
PosteriorDensity and the prior density values in
PriorDensity.
[
returns the figure handle of the figure containing the distributions.pointsUsed,posteriorDensity,priorDensity,FigureHandle]
= plot(___)
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Limitations
Because improper distributions (distributions with densities
that do not integrate to 1) are not well defined, plot
cannot plot them very well.
More About
Version History
Introduced in R2017a
See Also
Objects
conjugateblm|semiconjugateblm|diffuseblm|empiricalblm|customblm|mixconjugateblm|mixsemiconjugateblm|lassoblm