normalbvarm
Bayesian vector autoregression (VAR) model with normal conjugate prior and fixed covariance for data likelihood
Description
The Bayesian VAR model object normalbvarm specifies the prior distribution of the array of model coefficients Λ in an m-D VAR(p) model, where the innovations covariance matrix Σ is known and fixed. The prior distribution of Λ is the normal conjugate prior model.
In general, when you create a Bayesian VAR model object, it specifies the joint prior distribution and characteristics of the VARX model only. That is, the model object is a template intended for further use. Specifically, to incorporate data into the model for posterior distribution analysis, pass the model object and data to the appropriate object function.
Creation
Description
To create a normalbvarm object, use either the normalbvarm function (described here) or the bayesvarm function. The syntaxes for each function are similar, but the options differ. bayesvarm enables you to set prior hyperparameter values for Minnesota prior[1] regularization easily, whereas normalbvarm requires the entire specification of prior distribution hyperparameters.
PriorMdl = normalbvarm(numseries,numlags)numseries-D Bayesian VAR(numlags) model object PriorMdl, which specifies dimensionalities and prior assumptions for all model coefficients , where:
numseries= m, the number of response time series variables.numlags= p, the AR polynomial order.The prior distribution of λ is the normal conjugate prior model.
The fixed innovations covariance Σ is the m-by-m identity matrix.
PriorMdl = normalbvarm(numseries,numlags,Name,Value)NumSeries and P) using name-value pair arguments. Enclose each property name in quotes. For example, normalbvarm(3,2,'Sigma',4*eye(3),'SeriesNames',["UnemploymentRate" "CPI" "FEDFUNDS"]) specifies the names of the three response variables in the Bayesian VAR(2) model, and fixes the innovations covariance matrix at 4*eye(3).
Input Arguments
Number of time series m, specified as a positive integer. numseries specifies the dimensionality of the multivariate response variable yt and innovation εt.
numseries sets the NumSeries property.
Data Types: double
Number of lagged responses in each equation of yt, specified as a nonnegative integer. The resulting model is a VAR(numlags) model; each lag has a numseries-by-numseries coefficient matrix.
numlags sets the P property.
Data Types: double
Properties
You can set writable property values when you create the model object by using name-value argument syntax, or after you create the model object by using dot notation. For example, to create a 3-D Bayesian VAR(1) model and label the first through third response variables, and then include a linear time trend term, enter:
PriorMdl = normalbvarm(3,1,'SeriesNames',["UnemploymentRate" "CPI" "FEDFUNDS"]); PriorMdl.IncludeTrend = true;
Model Characteristics and Dimensionality
Model description, specified as a string scalar or character vector. The default value describes the model dimensionality, for example '2-Dimensional VAR(3) Model'.
Example: "Model 1"
Data Types: string | char
This property is read-only.
Number of time series m, specified as a positive integer. NumSeries specifies the dimensionality of the multivariate response variable yt and innovation εt.
Data Types: double
This property is read-only.
Multivariate autoregressive polynomial order, specified as a nonnegative integer. P is the maximum lag that has a nonzero coefficient matrix.
P specifies the number of presample observations required to initialize the model.
Data Types: double
Response series names, specified as a NumSeries length string vector. The default is ['Y1' 'Y2' ... 'Y. NumSeries']normalbvarm stores SeriesNames as a string vector.
Example: ["UnemploymentRate" "CPI" "FEDFUNDS"]
Data Types: string
Flag for including a model constant c, specified as a value in this table.
| Value | Description |
|---|---|
false | Response equations do not include a model constant. |
true | All response equations contain a model constant. |
Data Types: logical
Flag for including a linear time trend term δt, specified as a value in this table.
| Value | Description |
|---|---|
false | Response equations do not include a linear time trend term. |
true | All response equations contain a linear time trend term. |
Data Types: logical
Number of exogenous predictor variables in the model regression component, specified as a nonnegative integer. normalbvarm includes all predictor variables symmetrically in each response equation.
Distribution Hyperparameters
Mean of the multivariate normal prior on λ, specified as a NumSeries*-by-1 numeric vector, where kk = NumSeries*P + IncludeIntercept + IncludeTrend + NumPredictors
Mu(1: corresponds to all coefficients in the equation of response variable k)SeriesNames(1), Mu(( corresponds to all coefficients in the equation of response variable k + 1):(2*k))SeriesNames(2), and so on. For a set of indices corresponding to an equation:
Elements
1throughNumSeriescorrespond to the lag 1 AR coefficients of the response variables ordered bySeriesNames.Elements
NumSeries + 1through2*NumSeriescorrespond to the lag 2 AR coefficients of the response variables ordered bySeriesNames.In general, elements
(throughq– 1)*NumSeries + 1q*NumSeriesqSeriesNames.If
IncludeConstantistrue, elementNumSeries*P + 1is the model constant.If
IncludeTrendistrue, elementNumSeries*P + 2is the linear time trend coefficient.If
NumPredictors> 0, elementsNumSeries*P + 3throughk
This figure shows the structure of the transpose of Mu for a 2-D VAR(3) model that contains a constant vector and four exogenous predictors:
where
ϕq,jk is element (j,k) of the lag q AR coefficient matrix.
cj is the model constant in the equation of response variable j.
Bju is the regression coefficient of the exogenous variable u in the equation of response variable j.
Tip
bayesvarm enables you to specify Mu easily by using the Minnesota regularization method. To specify Mu directly:
Set separate variables for the prior mean of each coefficient matrix and vector.
Horizontally concatenate all coefficient means in this order:
Vectorize the transpose of the coefficient mean matrix.
Mu = Coeff.'; Mu = Mu(:);
Data Types: double
Conditional covariance matrix of multivariate normal prior on λ, specified as a NumSeries*-by-kNumSeries* symmetric, positive definite matrix, where kk = NumSeries*P + IncludeIntercept + IncludeTrend + NumPredictors
Row and column indices correspond to the model coefficients in the same way as Mu. For example, consider a 3-D VAR(2) model containing a constant and four exogenous variables.
V(1,1)is Var(ϕ1,11).V(5,6)is Cov(ϕ2,12,ϕ2,13).V(8,9)is Cov(β11,β12).
Tip
bayesvarm enables you to create any Bayesian VAR prior model and specify V easily by using the Minnesota regularization method.
Data Types: double
Fixed innovations covariance matrix Σ of the NumSeries innovations at each time t = 1,...,T, specified as a NumSeries-by-NumSeries positive definite numeric matrix. Rows and columns correspond to innovations in the equations of the response variables ordered by SeriesNames.
Sigma sets the Covariance property.
Data Types: double
VAR Model Parameters Derived from Distribution Hyperparameters
This property is read-only.
Distribution mean of the autoregressive coefficient matrices Φ1,…,Φp associated with the lagged responses, specified as a P-D cell vector of NumSeries-by-NumSeries numeric matrices.
AR{ is
Φj}j, the coefficient matrix of
lag j. Rows correspond to equations and columns correspond to
lagged response variables; SeriesNames determines the order of
response variables and equations. Coefficient signs are those of the VAR model expressed
in difference-equation notation.
If P = 0, AR is an empty cell. Otherwise, AR is the collection of AR coefficient means extracted from Mu.
Data Types: cell
This property is read-only.
Distribution mean of the model constant c (or intercept), specified as a NumSeries-by-1 numeric vector. Constant( is the constant in equation j)j; SeriesNames determines the order of equations.
If IncludeConstant = false, Constant is an empty array. Otherwise, Constant is the model constant vector mean extracted from Mu.
Data Types: double
This property is read-only.
Distribution mean of the linear time trend δ, specified as a NumSeries-by-1 numeric vector. Trend( is the linear time trend in equation j)j; SeriesNames determines the order of equations.
If IncludeTrend = false (the default), Trend is an empty array. Otherwise, Trend is the linear time trend coefficient mean extracted from Mu.
Data Types: double
This property is read-only.
Distribution mean of the regression coefficient matrix B associated with the exogenous predictor variables, specified as a NumSeries-by-NumPredictors numeric matrix.
Beta( contains the regression coefficients of each predictor in the equation of response variable j
yj,:)j,t. Beta(:, contains the regression coefficient in each equation of predictor xk. By default, all predictor variables are in the regression component of all response equations. You can down-weight a predictor from an equation by specifying, for the corresponding coefficient, a prior mean of 0 in k)Mu and a small variance in V.
When you create a model, the predictor variables are hypothetical. You specify predictor data when you operate on the model (for example, when you estimate the posterior by using estimate). Columns of the predictor data determine the order of the columns of Beta.
Data Types: double
This property is read-only.
Fixed innovations covariance matrix Σ of the NumSeries innovations at each time t = 1,...,T, specified as a NumSeries-by-NumSeries symmetric, positive definite numeric matrix. Rows and columns correspond to innovations in the equations of the response variables ordered by SeriesNames.
The Sigma property sets Covariance.
Data Types: double
Object Functions
estimate | Estimate posterior distribution of Bayesian vector autoregression (VAR) model parameters |
forecast | Forecast responses from Bayesian vector autoregression (VAR) model |
simsmooth | Simulation smoother of Bayesian vector autoregression (VAR) model |
simulate | Simulate coefficients and innovations covariance matrix of Bayesian vector autoregression (VAR) model |
summarize | Distribution summary statistics of Bayesian vector autoregression (VAR) model |
Examples
Consider the 3-D VAR(4) model for the US inflation (INFL), unemployment (UNRATE), and federal funds (FEDFUNDS) rates.
For all , is a series of independent 3-D normal innovations with a mean of 0 and fixed covariance = , the 3-D identity matrix. Assume that the prior distribution , where is a 39-by-1 vector of means and is the 39-by-39 covariance matrix.
Create a normal conjugate prior model for the 3-D VAR(4) model parameters.
numseries = 3; numlags = 4; PriorMdl = normalbvarm(numseries,numlags)
PriorMdl =
normalbvarm with properties:
Description: "3-Dimensional VAR(4) Model"
NumSeries: 3
P: 4
SeriesNames: ["Y1" "Y2" "Y3"]
IncludeConstant: 1
IncludeTrend: 0
NumPredictors: 0
Mu: [39×1 double]
V: [39×39 double]
Sigma: [3×3 double]
AR: {[3×3 double] [3×3 double] [3×3 double] [3×3 double]}
Constant: [3×1 double]
Trend: [3×0 double]
Beta: [3×0 double]
Covariance: [3×3 double]
PriorMdl is a normalbvarm Bayesian VAR model object representing the prior distribution of the coefficients of the 3-D VAR(4) model. The command line display shows properties of the model. You can display properties by using dot notation.
Display the prior mean matrices of the four AR coefficients by setting each matrix in the cell to a variable.
AR1 = PriorMdl.AR{1}AR1 = 3×3
0 0 0
0 0 0
0 0 0
AR2 = PriorMdl.AR{2}AR2 = 3×3
0 0 0
0 0 0
0 0 0
AR3 = PriorMdl.AR{3}AR3 = 3×3
0 0 0
0 0 0
0 0 0
AR4 = PriorMdl.AR{4}AR4 = 3×3
0 0 0
0 0 0
0 0 0
normalbvarm centers all AR coefficients at 0 by default. The AR property is read only, but it is derived from the writeable property Mu.
Display the fixed innovations covariance .
PriorMdl.Covariance
ans = 3×3
1 0 0
0 1 0
0 0 1
Covariance is a read-only property. To set the value , use the 'Sigma' name-value pair argument or specify the Sigma property by using dot notation. For example:
PriorMdl.Sigma = 4*eye(PriorMdl.NumSeries);
Consider the 3-D VAR(4) model of Create Normal Conjugate Prior Model.
Suppose econometric theory dictates that
Create a normal conjugate prior model for the VAR model coefficients. Specify the value of .
numseries = 3;
numlags = 4;
Sigma = [10e-5 0 10e-4; 0 0.1 -0.2; 10e-4 -0.2 1.6];
PriorMdl = normalbvarm(numseries,numlags,'Sigma',Sigma)PriorMdl =
normalbvarm with properties:
Description: "3-Dimensional VAR(4) Model"
NumSeries: 3
P: 4
SeriesNames: ["Y1" "Y2" "Y3"]
IncludeConstant: 1
IncludeTrend: 0
NumPredictors: 0
Mu: [39×1 double]
V: [39×39 double]
Sigma: [3×3 double]
AR: {[3×3 double] [3×3 double] [3×3 double] [3×3 double]}
Constant: [3×1 double]
Trend: [3×0 double]
Beta: [3×0 double]
Covariance: [3×3 double]
Because is fixed for normalbvarm prior models, PriorMdl.Sigma and PriorMdl.Covariance are equal.
PriorMdl.Sigma
ans = 3×3
0.0001 0 0.0010
0 0.1000 -0.2000
0.0010 -0.2000 1.6000
PriorMdl.Covariance
ans = 3×3
0.0001 0 0.0010
0 0.1000 -0.2000
0.0010 -0.2000 1.6000
Consider a 1-D Bayesian AR(2) model for the daily NASDAQ returns from January 2, 1990 through December 31, 2001.
The coefficient prior distribution is , where is a 3-by-1 vector of coefficient means and is a 3-by-3 covariance matrix. Assume Var() is 2.
Create a normal conjugate prior model for the AR(2) model parameters.
numseries = 1;
numlags = 2;
PriorMdl = normalbvarm(numseries,numlags,'Sigma',2)PriorMdl =
normalbvarm with properties:
Description: "1-Dimensional VAR(2) Model"
NumSeries: 1
P: 2
SeriesNames: "Y1"
IncludeConstant: 1
IncludeTrend: 0
NumPredictors: 0
Mu: [3×1 double]
V: [3×3 double]
Sigma: 2
AR: {[0] [0]}
Constant: 0
Trend: [1×0 double]
Beta: [1×0 double]
Covariance: 2
In the 3-D VAR(4) model of Create Normal Conjugate Prior Model, consider excluding lags 2 and 3 from the model.
You cannot exclude coefficient matrices from models, but you can specify high prior tightness on zero for coefficients that you want to exclude.
Create a normal conjugate prior model for the 3-D VAR(4) model parameters. Specify response variable names.
By default, AR coefficient prior means are zero. Specify high tightness values for lags 2 and 3 by setting their prior variances to 1e-6. Leave all other coefficient tightness values at their defaults:
1for AR coefficient variances1e3for constant vector variances0for all coefficient covariances
numseries = 3; numlags = 4; seriesnames = ["INFL"; "UNRATE"; "FEDFUNDS"]; vPhi1 = ones(numseries,numseries); vPhi2 = 1e-6*ones(numseries,numseries); vPhi3 = 1e-6*ones(numseries,numseries); vPhi4 = ones(numseries,numseries); vc = 1e3*ones(3,1); Vmat = [vPhi1 vPhi2 vPhi3 vPhi4 vc]'; V = diag(Vmat(:)); PriorMdl = normalbvarm(numseries,numlags,'SeriesNames',seriesnames,... 'V',V)
PriorMdl =
normalbvarm with properties:
Description: "3-Dimensional VAR(4) Model"
NumSeries: 3
P: 4
SeriesNames: ["INFL" "UNRATE" "FEDFUNDS"]
IncludeConstant: 1
IncludeTrend: 0
NumPredictors: 0
Mu: [39×1 double]
V: [39×39 double]
Sigma: [3×3 double]
AR: {[3×3 double] [3×3 double] [3×3 double] [3×3 double]}
Constant: [3×1 double]
Trend: [3×0 double]
Beta: [3×0 double]
Covariance: [3×3 double]
normalbvarm options enable you to specify coefficient prior hyperparameter values directly, but bayesvarm options are well suited for tuning hyperparameters following the Minnesota regularization method.
Consider the 3-D VAR(4) model of Create Normal Conjugate Prior Model. The model contains 39 coefficients. For coefficient sparsity, create a normal conjugate Bayesian VAR model by using bayesvarm. Specify the following, a priori:
Each response is an AR(1) model, on average, with lag 1 coefficient 0.75.
Prior self-lag coefficients have variance 100. This large variance setting allows the data to influence the posterior more than the prior.
Prior cross-lag coefficients have variance 1. This small variance setting tightens the cross-lag coefficients to zero during estimation.
Prior coefficient covariances decay with increasing lag at a rate of 2 (that is, lower lags are more important than higher lags).
The innovations covariance = .
numseries = 3; numlags = 4; seriesnames = ["INFL"; "UNRATE"; "FEDFUNDS"]; Sigma = eye(numseries); PriorMdl = bayesvarm(numseries,numlags,'ModelType','normal','Sigma',Sigma,... 'Center',0.75,'SelfLag',100,'CrossLag',1,'Decay',2,'SeriesNames',seriesnames)
PriorMdl =
normalbvarm with properties:
Description: "3-Dimensional VAR(4) Model"
NumSeries: 3
P: 4
SeriesNames: ["INFL" "UNRATE" "FEDFUNDS"]
IncludeConstant: 1
IncludeTrend: 0
NumPredictors: 0
Mu: [39×1 double]
V: [39×39 double]
Sigma: [3×3 double]
AR: {[3×3 double] [3×3 double] [3×3 double] [3×3 double]}
Constant: [3×1 double]
Trend: [3×0 double]
Beta: [3×0 double]
Covariance: [3×3 double]
Display all prior coefficient means.
Phi1 = PriorMdl.AR{1}Phi1 = 3×3
0.7500 0 0
0 0.7500 0
0 0 0.7500
Phi2 = PriorMdl.AR{2}Phi2 = 3×3
0 0 0
0 0 0
0 0 0
Phi3 = PriorMdl.AR{3}Phi3 = 3×3
0 0 0
0 0 0
0 0 0
Phi4 = PriorMdl.AR{4}Phi4 = 3×3
0 0 0
0 0 0
0 0 0
Display a heatmap of the prior coefficient covariances for each response equation.
numexocoeffseqn = PriorMdl.IncludeConstant + ... PriorMdl.IncludeTrend + PriorMdl.NumPredictors; % Number of exogenous coefficients per equation numcoeffseqn = PriorMdl.NumSeries*PriorMdl.P + numexocoeffseqn; % Total number of coefficients per equation arcoeffnames = strings(numseries,numlags,numseries); for j = 1:numseries % Equations for r = 1:numlags for k = 1:numseries % Response Variables arcoeffnames(k,r,j) = "\phi_{"+r+","+j+k+"}"; end end arcoeffseqn = arcoeffnames(:,:,j); idx = ((j-1)*numcoeffseqn + 1):(numcoeffseqn*j) - numexocoeffseqn; Veqn = PriorMdl.V(idx,idx); figure heatmap(arcoeffseqn(:),arcoeffseqn(:),Veqn); title(sprintf('Equation of %s',seriesnames(j))) end
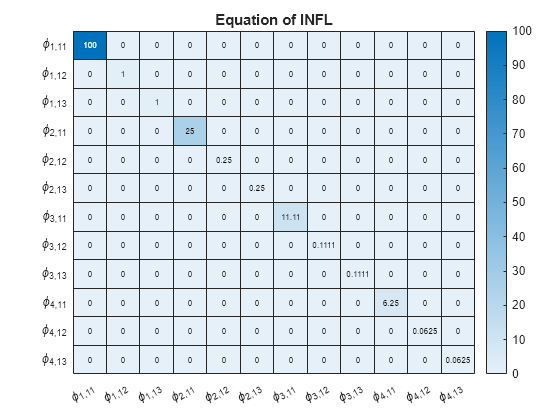
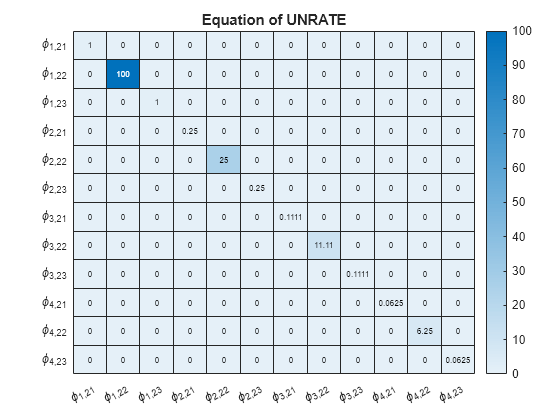
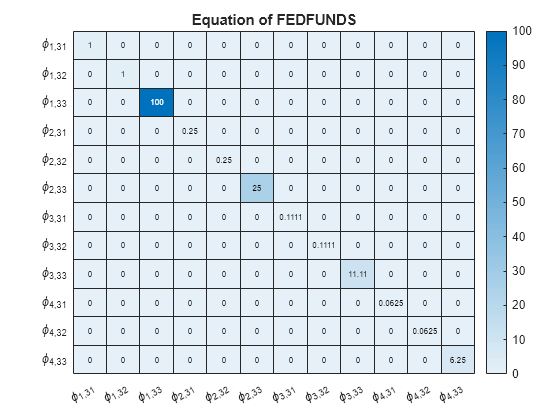
Consider the 3-D VAR(4) model of Create Normal Conjugate Prior Model. Estimate the posterior distribution, and generate forecasts from the corresponding posterior predictive distribution.
Load and Preprocess Data
Load the US macroeconomic data set. Compute the inflation rate. Plot all response series.
load Data_USEconModel seriesnames = ["INFL" "UNRATE" "FEDFUNDS"]; DataTimeTable.INFL = 100*[NaN; price2ret(DataTimeTable.CPIAUCSL)]; figure plot(DataTimeTable.Time,DataTimeTable{:,seriesnames}) legend(seriesnames)
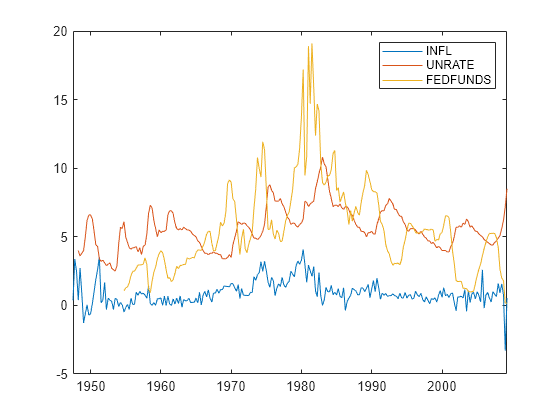
Stabilize the unemployment and federal funds rates by applying the first difference to each series.
DataTimeTable.DUNRATE = [NaN; diff(DataTimeTable.UNRATE)];
DataTimeTable.DFEDFUNDS = [NaN; diff(DataTimeTable.FEDFUNDS)];
seriesnames(2:3) = "D" + seriesnames(2:3);Remove all missing values from the data.
rmDataTimeTable = rmmissing(DataTimeTable);
Create Prior Model
Create a normal conjugate Bayesian VAR(4) prior model for the three response series. Specify the response variable names. Assume that the innovations covariance is the identity matrix.
numseries = numel(seriesnames);
numlags = 4;
PriorMdl = normalbvarm(numseries,numlags,'SeriesNames',seriesnames);Estimate Posterior Distribution
Estimate the posterior distribution by passing the prior model and entire data series to estimate.
PosteriorMdl = estimate(PriorMdl,rmDataTimeTable{:,seriesnames},'Display','equation');Bayesian VAR under normal priors and fixed Sigma
Effective Sample Size: 197
Number of equations: 3
Number of estimated Parameters: 39
VAR Equations
| INFL(-1) DUNRATE(-1) DFEDFUNDS(-1) INFL(-2) DUNRATE(-2) DFEDFUNDS(-2) INFL(-3) DUNRATE(-3) DFEDFUNDS(-3) INFL(-4) DUNRATE(-4) DFEDFUNDS(-4) Constant
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
INFL | 0.1260 -0.4400 0.1049 0.3176 -0.0545 0.0440 0.4173 0.2421 0.0515 0.0247 -0.1639 0.0080 0.1064
| (0.1367) (0.2673) (0.0700) (0.1551) (0.2854) (0.0739) (0.1536) (0.2811) (0.0766) (0.1605) (0.2652) (0.0708) (0.1483)
DUNRATE | -0.0236 0.4440 0.0350 0.0900 0.2295 0.0520 -0.0330 0.0567 0.0010 0.0298 -0.1665 0.0104 -0.0536
| (0.1367) (0.2673) (0.0700) (0.1551) (0.2854) (0.0739) (0.1536) (0.2811) (0.0766) (0.1605) (0.2652) (0.0708) (0.1483)
DFEDFUNDS | -0.1514 -1.3408 -0.2762 0.3275 -0.2971 -0.3041 0.2609 -0.6971 0.0130 -0.0692 0.1392 -0.1341 -0.3902
| (0.1367) (0.2673) (0.0700) (0.1551) (0.2854) (0.0739) (0.1536) (0.2811) (0.0766) (0.1605) (0.2652) (0.0708) (0.1483)
Innovations Covariance Matrix
| INFL DUNRATE DFEDFUNDS
--------------------------------------
INFL | 1 0 0
| (0) (0) (0)
DUNRATE | 0 1 0
| (0) (0) (0)
DFEDFUNDS | 0 0 1
| (0) (0) (0)
Because the prior is conjugate for the data likelihood, the posterior is analytically tractable. By default, estimate uses the first four observations as a presample to initialize the model.
Generate Forecasts from Posterior Predictive Distribution
From the posterior predictive distribution, generate forecasts over a two-year horizon. Because sampling from the posterior predictive distribution requires the entire data set, specify the prior model in forecast instead of the posterior.
fh = 8;
rng(1); % For reproducibility
FY = forecast(PriorMdl,fh,rmDataTimeTable{:,seriesnames});FY is an 8-by-3 matrix of forecasts.
Plot the end of the data set and the forecasts.
fp = rmDataTimeTable.Time(end) + calquarters(1:fh);
figure
plotdata = [rmDataTimeTable{end - 10:end,seriesnames}; FY];
plot([rmDataTimeTable.Time(end - 10:end); fp'],plotdata)
hold on
plot([fp(1) fp(1)],ylim,'k-.')
legend(seriesnames)
title('Data and Forecasts')
hold off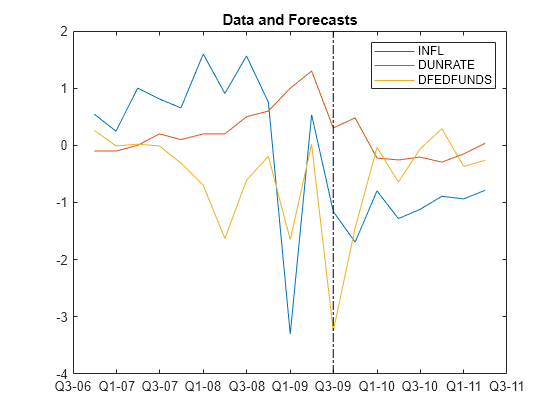
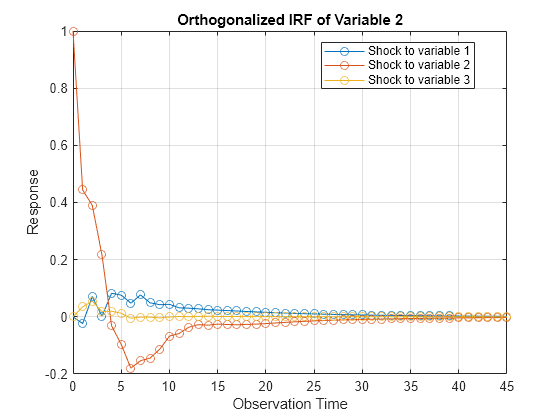
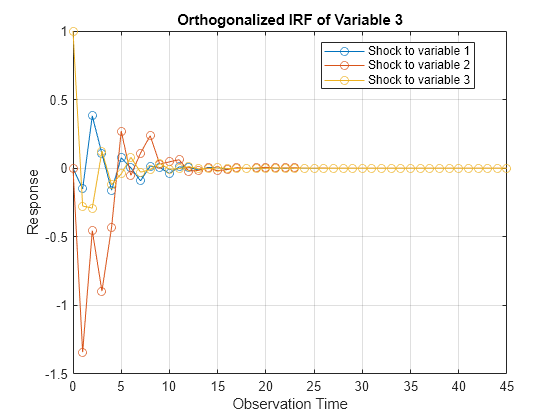
Compute Impulse Responses
Plot impulse response functions by passing posterior estimations to armairf.
armairf(PosteriorMdl.AR,[],'InnovCov',PosteriorMdl.Covariance)
More About
A Bayesian VAR model treats all coefficients and the innovations covariance matrix as random variables in the m-dimensional, stationary VARX(p) model. The model has one of the three forms described in this table.
| Model | Equation |
|---|---|
| Reduced-form VAR(p) in difference-equation notation |
|
| Multivariate regression |
|
| Matrix regression |
|
For each time t = 1,...,T:
yt is the m-dimensional observed response vector, where m =
numseries.Φ1,…,Φp are the m-by-m AR coefficient matrices of lags 1 through p, where p =
numlags.c is the m-by-1 vector of model constants if
IncludeConstantistrue.δ is the m-by-1 vector of linear time trend coefficients if
IncludeTrendistrue.Β is the m-by-r matrix of regression coefficients of the r-by-1 vector of observed exogenous predictors xt, where r =
NumPredictors. All predictor variables appear in each equation.which is a 1-by-(mp + r + 2) vector, and Zt is the m-by-m(mp + r + 2) block diagonal matrix
where 0z is a 1-by-(mp + r + 2) vector of zeros.
, which is an (mp + r + 2)-by-m random matrix of the coefficients, and the m(mp + r + 2)-by-1 vector λ = vec(Λ).
εt is an m-by-1 vector of random, serially uncorrelated, multivariate normal innovations with the zero vector for the mean and the m-by-m matrix Σ for the covariance. This assumption implies that the data likelihood is
where f is the m-dimensional multivariate normal density with mean ztΛ and covariance Σ, evaluated at yt.
Before considering the data, you impose a joint prior distribution assumption on (Λ,Σ), which is governed by the distribution π(Λ,Σ). In a Bayesian analysis, the distribution of the parameters is updated with information about the parameters obtained from the data likelihood. The result is the joint posterior distribution π(Λ,Σ|Y,X,Y0), where:
Y is a T-by-m matrix containing the entire response series {yt}, t = 1,…,T.
X is a T-by-m matrix containing the entire exogenous series {xt}, t = 1,…,T.
Y0 is a p-by-m matrix of presample data used to initialize the VAR model for estimation.
The normal conjugate prior model, outlined in [1], is an m-D Bayesian VAR model in which the innovations covariance matrix Σ is known and fixed, while the coefficient vector λ = vec(Λ) has the prior distribution
where:
r =
NumPredictors.1c is 1 if
IncludeConstantis true, and 0 otherwise.1δ is 1 if
IncludeTrendis true, and 0 otherwise.
The posterior distribution is proper and analytically tractable.
where:
References
[1] Litterman, Robert B. "Forecasting with Bayesian Vector Autoregressions: Five Years of Experience." Journal of Business and Economic Statistics 4, no. 1 (January 1986): 25–38. https://doi.org/10.2307/1391384.
Version History
Introduced in R2020a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)