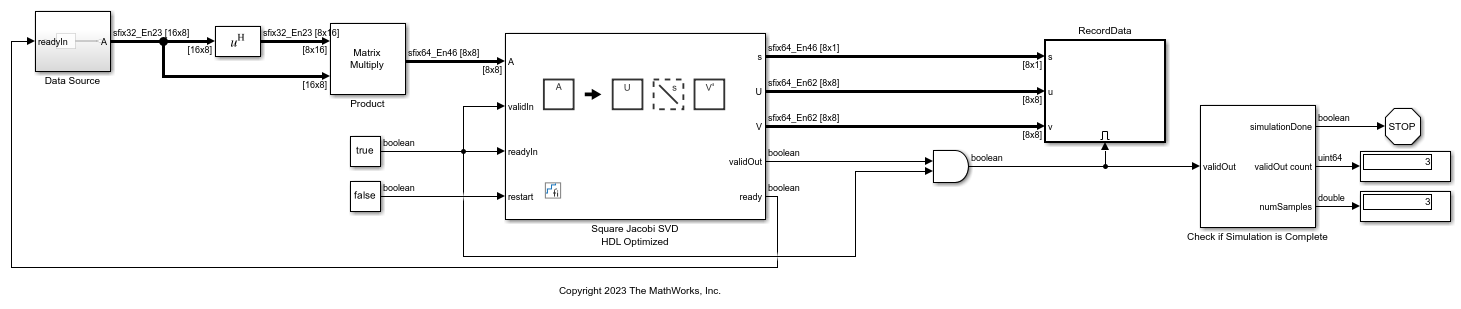
Non-Square Jacobi SVD HDL Optimized
Libraries:
Fixed-Point Designer HDL Support /
Matrices and Linear Algebra /
Matrix Factorizations
Description
Use the Non-Square Jacobi SVD HDL Optimized block to perform singular value
decomposition (SVD) on non-square matrices using QR decomposition and the two-sided Jacobi
algorithm. This block consists of a Real Partial-Systolic QR
Decomposition or Complex Partial-Systolic
QR Decomposition block, depending on your configuration, and a Square Jacobi SVD HDL
Optimized. Given a matrix A with more rows than columns, the
Non-Square Jacobi SVD HDL Optimized block uses QR decomposition to preprocess
the input, then uses the two-sided Jacobi method to produce a vector s of
nonnegative elements and unitary matrices U and V such
that A =
U*diag(s)*V'.
Note
For square matrices, use the Square Jacobi SVD HDL Optimized block.
Examples
Extended Examples
Limitations
To optimize HDL efficiency, if the input matrix A is not full rank, then the output U matrix is orthonormal up to the rank of A. is still valid. V is always orthonormal regardless of the rank of A.
For example, if , then A has rank 2 and .
If the input matrix is full rank, then , where I = eye(n).
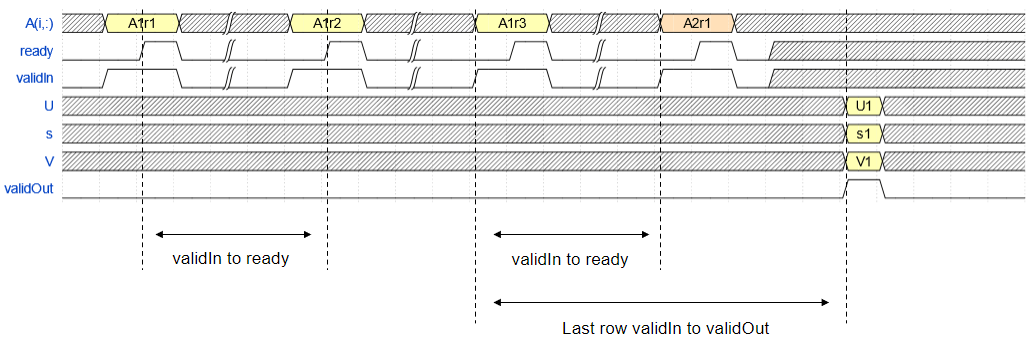
Ports
Input
Output
Parameters
Tips
The behavior of the Non-Square Jacobi SVD HDL Optimized block is equivalent to
[U,s,V] = fixed.jacobiSVD(A)whenAis a non-square matrix. If the input data type is fixed point with binary-point scaling, the function and the block provide bit-exact results. However, if the input data type is floating point, small numerical differences may exist between the function and the block.The Number of Jacobi iterations block parameter is equivalent to the
numberOfSweepsinput argument for thefixed.jacobiSVDfunction.
Algorithms
References
[1] Arm Developer. "AMBA AXI and ACE Protocol Specification Version E." https://developer.arm.com/documentation/ihi0022/e/AMBA-AXI3-and-AXI4-Protocol-Specification/Single-Interface-Requirements/Basic-read-and-write-transactions/Handshake-process.
[2] Jacobi, Carl G. J., "Über ein leichtes Verfahren die in der Theorie der Säcularstörungen vorkommenden Gleichungen numerisch aufzulösen." Journal fur die reine und angewandte Mathematik 30 (1846): 51–94.
[3] Forsythe, George E. and Peter Henrici. "The Cyclic Jacobi Method for Computing the Principal Values of a Complex Matrix." Transactions of the American Mathematical Society 94, no. 1 (January 1960): 1-23.
[4] Shiri, Aidin and Ghader Khosroshahi. "An FPGA Implementation of Singular Value Decomposition", ICEE 2019: 27th Iranian Conference on Electrical Engineering, Yazd, Iran, April 30–May 2, 2019, 416-22, IEEE.
[5] Golub, Gene H. and Charles F. Van Loan. Matrix Computations, 4th ed. Baltimore, MD: Johns Hopkins University Press, 2013.
[6] Athi, Mrudula V., Seyed R. Zekavat, and Alan A. Struthers. "Real-Time Signal Processing of Massive Sensor Arrays via a Parallel Fast Converging SVD Algorithm: Latency, Throughput, and Resource Analysis." IEEE Sensors Journal 16, no. 18 (January 2016): 2519-26. https://doi.org/10.1109/JSEN.2016.2517040.
[7] Brent, Richard P., Franklin T. Luk, and Charles Van Loan. "Computation of the Singular Value Decomposition Using Mesh-Connected Processors." Journal of VLSI and Computer Systems 1, 3 (1985): 242–70.
[8] Hemkumar, Nariankadu D. A Systolic VLSI Architecture for Complex SVD. Master’s thesis, Rice University, 1991.
[9] Duryea, R. A. Finite Precision Arithmetic in Singular Value Decomposition Architectures. Ph.D. thesis, Cornell University, 1987.
[10] Cavallaro, Joseph R. and Franklin T. Luk. 1987. "CORDIC Arithmetic for an SVD Processor." 1987 IEEE 8th Symposium on Computer Arithmetic (ARITH), Como, Italy, May 18-21, 1987, 113-20. IEEE. https://doi.org/10.1109/ARITH.1987.6158686.
Extended Capabilities
Version History
Introduced in R2023b