bigimageshow
Display 2-D blockedImage object
Description
Note
imageshow is
recommended over bigimageshow because it provides faster performance,
improved rendering quality, and a simpler interface. (since R2024b)
A bigimageshow object displays data from a
blockedImage object. The bigimageshow object
progressively loads image data based on image extents and screen resolution.
Creation
Description
bigimageshow( displays the 2-D blocked
image bim)bim.
For categorical data, bigimageshow sets the axis colormap to
parula. For numeric data, gray is the default
colormap.
b = bigimageshow(___) returns the
bigimageshow object b. Use b to
modify the display settings after you display the blocked image.
b = bigimageshow(___,Name=Value)
For example, bigimageshow(bim,GridVisible="on",GridLineStyle=":")
displays the blocked image, bim, and overlays dotted grid lines.
Input Arguments
Blocked image, specified as a blockedImage object.
Parent axes of bigimageshow object, specified as an axes
object.
Properties
Parent axes of the bigimageshow object, specified as an
axes object. If you do not specify a parent,
bigimageshow uses the handle to the current figure,
gca. If a figure does not exist, bigimageshow
creates a new figure.
2-D blockedImage object to display, specified as a blockedImage
object.
Color data mapping method, specified as "direct" or
"scaled". Use this property to control the mapping of color data
values in CData into the colormap. CData must be
a vector or a matrix defining indexed colors. This property has no effect if
CData is a 3-D array defining RGB colors.
The methods have these effects:
"direct"— Interpret the values as indices into the current colormap. Values with a decimal portion are fixed to the nearest lower integer.If the values are of type
doubleorsingle, values of1or less map to the first color in the colormap. Values equal to or greater than the length of the colormap map to the last color in the colormap.If the values are of type
uint8,uint16,uint32,uint64,int8,int16,int32, orint64, values of0or less map to the first color in the colormap. Values equal to or greater than the length of the colormap map to the last color in the colormap (or up to the range limits of the type).If the values are of type
logical, values of0map to the first color in the colormap and values of1map to the second color in the colormap.
"scaled"— Scale the values to range between the minimum and maximum color limits. TheCLimproperty of the axes contains the color limits.
Transparency data, specified in one of these forms:
Numeric scalar — Use a consistent transparency across the entire image.
2-D
blockedImageobject — Transparency data must have the same rows and columns extent as theCData2-DblockedImageobject. The blocked image can have multiple resolution levels, in which case,bigimageshowselects the level closest to the currentResolutionLevelfor display.
The AlphaDataMapping property controls how MATLAB® interprets the alpha data transparency values.
Example: 0.5
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | logical
Interpretation of AlphaData values, specified as one of these
values:
"none"— Interpret the values as transparency values. A value of 1 or greater is completely opaque, a value of 0 or less is completely transparent, and a value between 0 and 1 is semitransparent."scaled"— Map the values into the figure’s alphamap. The minimum and maximum alpha limits of the axes determine the alpha data values that map to the first and last elements in the alphamap, respectively. For example, if the alpha limits are[3 5], alpha data values less than or equal to3map to the first element in the alphamap. Alpha data values greater than or equal to5map to the last element in the alphamap. TheALimproperty of the axes contains the alpha limits. TheAlphamapproperty of the figure contains the alphamap."direct"— Interpret the values as indices into the figure’s alphamap. Values with a decimal portion are fixed to the nearest lower integer:If the values are of type
doubleorsingle, values of 1 or less map to the first element in the alphamap. Values equal to or greater than the length of the alphamap map to the last element in the alphamap.If the values are of type integer, then values of 0 or less map to the first element in the alphamap. Values equal to or greater than the length of the alphamap map to the last element in the alphamap (or up to the range limits of the type). The integer types are
uint8,uint16,uint32,uint64,int8,int16,int32, andint64.If the values are of type
logical, values of0map to the first element in the alphamap and values of1map to the second element in the alphamap.
Resolution level of the 2-D blockedImage object to display,
specified as a positive integer that identifies a resolution level of the 2-D
blockedImage object in the CData property.
Resolution level can also be specified as "fine" or
"coarse" corresponding to these two limits. The default value is
computed based on available screen space and resolution.
Selection mode for resolution level, specified as one of these values:
"auto"— Automatically select resolution level based on parent axes and available screen size."manual"— Manually specify resolution level by setting theResolutionLevelproperty.
Grid visibility, specified as "off" or "on".
bigimageshow spaces the grid in world units to include as many
pixels as specified by CData.BlockSize at the
current GridResolutionLevel.
Resolution level of blocked image at which to show grid, specified as one of these values:
positive integer — Display the grid specified as a numeric scalar that identifies a resolution level of the 2-D
blockedImageobject inCDataproperty. Value is between 1 and the value of theNumLevelsproperty of the blocked image in thebigimageshowCDataproperty."fine"— Display the grid at the finest resolution level."coarse"— Display the grid at the coarsest resolution level.
By default, GridLevel has the same value as
ResolutionLevel property.
Selection mode for grid level, specified as one of these values:
"auto"— Select the grid resolution level to match the image data resolution levelResolutionLevel."manual"— Manually specify the grid resolution level by setting theGridLevelproperty.
Grid line color, specified as an RGB triplet, a hexadecimal color code, a color
name, or a short color name. To display the grid lines, set the
GridVisible property to "on".
For a custom color, specify an RGB triplet or a hexadecimal color code.
An RGB triplet is a three-element row vector whose elements specify the intensities of the red, green, and blue components of the color. The intensities must be in the range
[0,1], for example,[0.4 0.6 0.7].A hexadecimal color code is a string scalar or character vector that starts with a hash symbol (
#) followed by three or six hexadecimal digits, which can range from0toF. The values are not case sensitive. Therefore, the color codes"#FF8800","#ff8800","#F80", and"#f80"are equivalent.
Alternatively, you can specify some common colors by name. This table lists the named color options, the equivalent RGB triplets, and the hexadecimal color codes.
| Color Name | Short Name | RGB Triplet | Hexadecimal Color Code | Appearance |
|---|---|---|---|---|
"red" | "r" | [1 0 0] | "#FF0000" |
|
"green" | "g" | [0 1 0] | "#00FF00" |
|
"blue" | "b" | [0 0 1] | "#0000FF" |
|
"cyan"
| "c" | [0 1 1] | "#00FFFF" |
|
"magenta" | "m" | [1 0 1] | "#FF00FF" |
|
"yellow" | "y" | [1 1 0] | "#FFFF00" |
|
"black" | "k" | [0 0 0] | "#000000" |
|
"white" | "w" | [1 1 1] | "#FFFFFF" |
|
This table lists the default color palettes for plots in the light and dark themes.
| Palette | Palette Colors |
|---|---|
Before R2025a: Most plots use these colors by default. |
|
|
|
You can get the RGB triplets and hexadecimal color codes for these palettes using the orderedcolors and rgb2hex functions. For example, get the RGB triplets for the "gem" palette and convert them to hexadecimal color codes.
RGB = orderedcolors("gem");
H = rgb2hex(RGB);Before R2023b: Get the RGB triplets using RGB =
get(groot,"FactoryAxesColorOrder").
Before R2024a: Get the hexadecimal color codes using H =
compose("#%02X%02X%02X",round(RGB*255)).
Example: GridColor = [1 0 0]
Example: GridColor = "r"
Example: GridColor = "red"
Example: GridColor = "#FF0000"
Grid line transparency, specified as a number in the range [0, 1]. A value of
1 means completely opaque and a value of 0 means
completely transparent. To display the grid lines, set the
GridVisible property to "on".
Grid line width, specified as a positive number, measured in points. To display the
grid lines, set the GridVisible property to
"on".
Grid line style, specified as one of the line styles in this table.
| Line Style | Description | Resulting Line |
|---|---|---|
"-" | Solid line |
|
"--" | Dashed line |
|
":" | Dotted line |
|
"-." | Dash-dot line |
|
To display the grid lines, set the GridVisible
property to "on".
Note
The Interpolation
property no longer has an effect. For more information, see Version History. (since R2025a)
Interpolation method used to resample pixels, specified as
"linear" for bilinear interpolation, or
"nearest" for nearest neighbor interpolation.
Control image visibility, specified as one of these values:
"on"— Display thebigimageshowobject."off"— Hide the object without deleting it. You still can access the properties of an invisible object.
Object Functions
showmask | Show mask overlay at specified inclusion threshold |
hidemask | Hide mask overlay in bigimageshow object |
showlabels | Display label overlay on bigimageshow object |
hidelabels | Hide label overlay on bigimageshow object |
Examples
This example uses a modified version of a training image of a lymph node containing tumor tissue (tumor_091.tif) from the CAMELYON16 data set. The modified image has three coarse resolution levels and has been adjusted to enforce a consistent aspect ratio and to register features at each level.
Create a blocked image from the sample image.
bim = blockedImage("tumor_091R.tif");
Display the blocked image.
h = bigimageshow(bim);

Zoom in on a region in the image.
xlim([2100 2600]) ylim([1800 2300])

To view the image at the three resolution levels, specify a new value for the ResolutionLevel property. As you view each resolution level, note that the axis limits remain the same, but bigimageshow ensures that the images from other levels are correctly sized. When you set ResolutionLevel, the ResolutionLevelMode value changes to 'manual' automatically.
h.ResolutionLevel = 3; pause(1); h.ResolutionLevel = 2; pause(1); h.ResolutionLevel = 1; pause(1);

Create a blocked image from the sample image tumor_091R.tif. This sample image is a training image of a lymph node containing tumor tissue from the CAMELYON16 data set. The image has been modified to have three coarse resolution levels, and has been adjusted to enforce a consistent aspect ratio and to register features at each level.
bim = blockedImage('tumor_091R.tif','BlockSize', [128 128]);
Display the blocked image with bigimageshow. Specify that you want the grid to be visible at the finest resolution level (level 1). Also specify the color, width, and transparency of the grid lines.
h = bigimageshow(bim,... 'GridVisible','on','GridLevel',1,... 'GridLineWidth', 2,'GridColor','k','GridAlpha',0.3);

Create a blocked image from the sample image tumor_091R.tif. This sample image is a training image of a lymph node containing tumor tissue from the CAMELYON16 data set. The image has been modified to have three coarse resolution levels, and has been adjusted to enforce a consistent aspect ratio and to register features at each level.
bim = blockedImage("tumor_091R.tif");
Create a coarse mask using the blockedImage apply object function.
bmask = apply(bim, @(bs)im2gray(bs.Data)<120,Level=3);
Overlay the mask as an alpha layer.
ha1 = subplot(1,2,1); h = bigimageshow(bim); h.AlphaData = bmask; h.AlphaDataMapping = "direct"; alphamap([0.4 1]) h.Parent.Color = "r";

Independently visualize the mask.
ha2 = subplot(1,2,2);
bigimageshow(bmask);
linkaxes([ha1,ha2]);
%

Create a blocked image from the sample image tumor_091R.tif. This sample image is a training image of a lymph node containing tumor tissue from the CAMELYON16 data set. The image has been modified to have three coarse resolution levels, and has been adjusted to enforce a consistent aspect ratio and to register features at each level.
bim = blockedImage("tumor_091R.tif");
Create a mask using the coarsest resolution level of the blocked image.
bmask = apply(bim,@(im)im2gray(im.Data)<120,Level=3);
Display the blocked image with the mask.
h = bigimageshow(bim); showmask(h,bmask);

Experiment with different inclusion thresholds to get a better fit of the mask over the stained area. By default, the inclusion threshold is 0.5 and the block size is 625-by-670 pixels.
showmask(h,bmask,InclusionThreshold=0.2); showmask(h,bmask,InclusionThreshold=0); showmask(h,bmask,InclusionThreshold=0.06);

For two of the inclusion thresholds, experiment with a smaller block sizes to get a better fit of the mask over the stained area.
showmask(h,bmask,InclusionThreshold=0.06,BlockSize=[256 256]); showmask(h,bmask,InclusionThreshold=0.14,BlockSize=[256 256]);

When you are satisfied with the mask, use it to segment the lymph node.
bls = selectBlockLocations(bim,BlockSize=[256 256], ... Mask=bmask,InclusionThreshold=0.14); bregion = apply(bim, @(bs)bs.Data,BlockLocationSet=bls); figure bigimageshow(bregion); %

Create a blocked image from the sample image tumor_091R.tif. This sample image is a training image of a lymph node containing tumor tissue from the CAMELYON16 data set. The image has been modified to have three coarse resolution levels, and has been adjusted to enforce a consistent aspect ratio and to register features at each level.
bim = blockedImage("tumor_091R.tif");Create a label image at a coarse resolution level.
First get a single-resolution image. By default, gather gets data from the coarsest resolution level.
cim = gather(bim);
Convert the image to grayscale. Use multithresh to calculate three threshold values to convert the image into a four-level image.
cgim = im2gray(cim); numClasses = 4; thresh = multithresh(cgim,numClasses-1);
Segment the image into four regions using imquantize, specifying the threshold levels returned by multithresh.
labels = imquantize(cgim,thresh); imagesc(labels) axis square title("Coarse Label Image")

Convert the labels image back to a blockedImage object, using the same spatial referencing as the original image at the coarsest resolution level.
blabels = blockedImage(labels,WorldStart=bim.WorldStart(3,1:2),...
WorldEnd=bim.WorldEnd(3,1:2));Display the original blocked image.
figure hB = bigimageshow(bim);
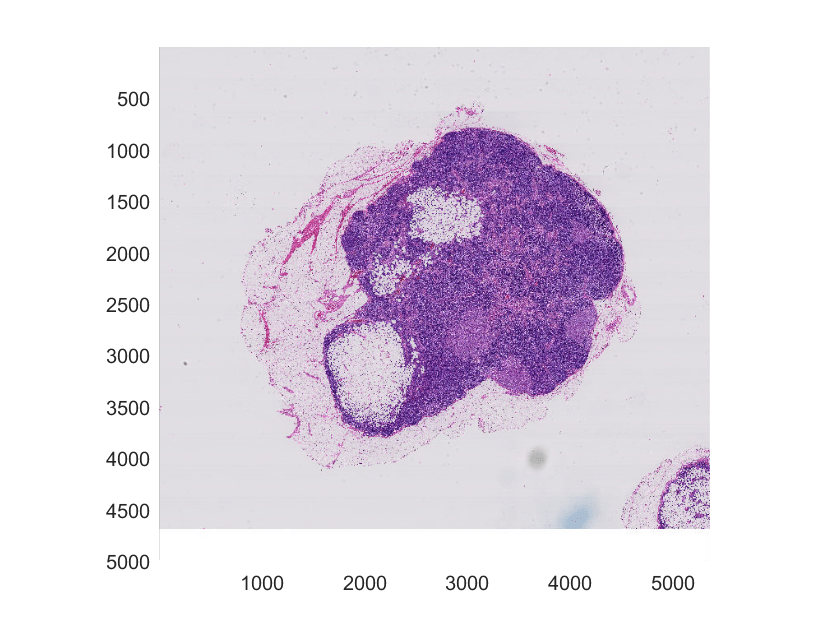
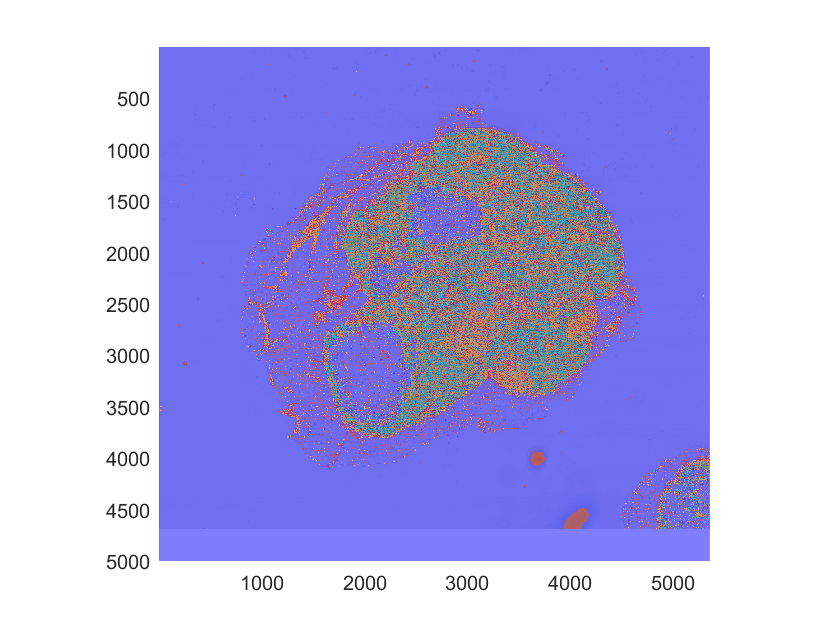
Overlay the labels image on the original blocked image.
showlabels(hB,blabels)
More About
To receive notification from the
bigimageshow object when certain events happen, set up
listeners for these events. You can specify a callback function
that executes when one of these events occurs. When the bigimageshow object
notifies your application through the listener, it returns data specific to the event. Look
at the event class for the specific event to see what is returned.
| Event Name | Trigger | Event Data | Event Attributes |
|---|---|---|---|
DataReadStarted (since R2023a) | The | event.EventData |
|
DataReadFinished (since R2023a) | The | event.EventData |
|
For example, this code adds listeners that change the title of the parent figure. The
title displays "Reading image data" while you zoom. The title displays
"Resolution level: " with the current resolution level after you
complete zooming.
bim = blockedImage("tumor_091R.tif"); h = bigimageshow(bim); listenerStarted = addlistener(h,"DataReadStarted", ... @(h,~)title("Reading image data",Parent=h.Parent)); listenerFinished = addlistener(h,"DataReadFinished", ... @(h,~)title("Resolution level: "+num2str(h.ResolutionLevel),Parent=h.Parent));
References
[1] Bejnordi, Babak Ehteshami, Mitko Veta, Paul Johannes van Diest, Bram van Ginneken, Nico Karssemeijer, Geert Litjens, Jeroen A. W. M. van der Laak, et al. “Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer.” JAMA 318, no. 22 (December 12, 2017): 2199–2210. https://doi.org/10.1001/jama.2017.14585.
[2] Grand Challenge. https://camelyon17.grand-challenge.org/Data/.
Version History
Introduced in R2019bThe Interpolation property no longer has an effect. All images are
displayed using nearest neighbor interpolation. To display blocked images using nearest
neighbor or bilinear interpolation, use the imageshow (since R2024b) function, which is recommended over
bigimageshow because it provides faster performance, improved
rendering quality, and a simpler interface.
The bigimageshow function now supports
DataReadStarted and DataReadFinished events.
The bigimageshow function now accepts blockedImage
objects as input for display.
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)