对不同量级的数据进行归一化
此示例说明如何通过归一化来用 griddata 改善散点数据插值结果。在某些情况下,归一化可以改善插值结果,但在其他情况下它可能会影响解的精确度。是否使用归一化需要根据插值数据的性质做出判断。
优势:归一化数据在自变量具有不同单位和迥异的规模时可能会改善插值结果。在这种情况下,缩放输入以使其具有类似的量级可以改善插值的数值形态。下面举例说明归一化的好处。假设
x表示从 500 到 3500 RPM 的引擎速度,y表示从 0 到 1 的引擎负载。x和y的规模相差几个数量级,并且它们具有不同单位。注意:如果各自变量具有相同的单位,则在归一化数据时应小心,即使各变量的规模不同也是如此。对于相同单位的数据,归一化会因添加方向偏差而影响基本三角剖分并最终降低插值的准确度,从而使解失真。例如,如果
x和y都表示位置并且都以“米”为单位,则进行归一化是错误的。此时不建议对x和y进行不一致的缩放,因为正东 10 米和正北 10 米在空间距离上是相同的。
创建一些样本数据,其中 y 中的值比 x 中的值大若干个数量级。假设 x 和 y 具有不同单位。
x = rand(1,500)/100; y = 2.*(rand(1,500)-0.5).*90; z = (x.*1e2).^2;
使用以上样本数据构建一个查询点网格。基于网格上的样本数据进行插值并绘制结果。
X = linspace(min(x),max(x),25); Y = linspace(min(y),max(y),25); [xq, yq] = meshgrid(X,Y); zq = griddata(x,y,z,xq,yq); plot3(x,y,z,'mo') hold on mesh(xq,yq,zq) xlabel('x') ylabel('y') hold off
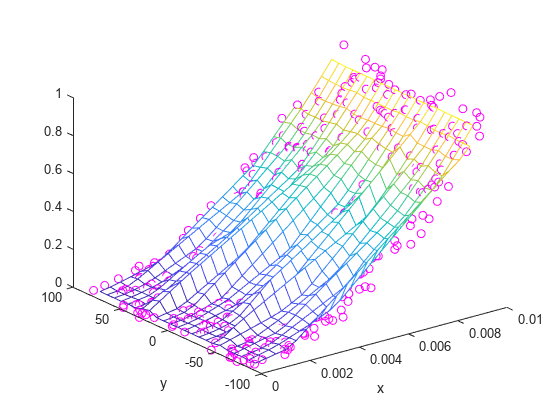
griddata 生成的结果不是很平滑,似乎存在含噪数据。这是由于自变量具有不同尺度导致的,因为一个变量的大小细微变化会导致另一个变量的大小发生巨大变化。
由于 x 和 y 具有不同单位,对它们进行归一化使其具有类似的量级,应该有助于产生更好的结果。使用 Z 分数对样本点进行归一化,并使用 griddata 重新生成插值。
% Normalize Sample Points x = normalize(x); y = normalize(y); % Regenerate Grid X = linspace(min(x),max(x),25); Y = linspace(min(y),max(y),25); [xq, yq] = meshgrid(X,Y); % Interpolate and Plot zq = griddata(x,y,z,xq,yq); plot3(x,y,z,'mo') hold on mesh(xq,yq,zq)

在这种情况下,对样本点进行归一化会使 griddata 计算出更平滑的解。