selectFeatures
Description
F = selectFeatures(R,featureNames)featureNames for the radiomics object
R.
Examples
Load an X-ray image into the workspace as a medicalImage object. Visualize the image.
data = medicalImage("forearmXrayImage1.dcm");
I = data.Pixels;
figure
imshow(I,[])Draw two regions of interest (ROI) in the X-ray image. Create masks from the ROIs.
roi1 = drawassisted(Color="g"); roi2 = drawassisted(Color="r");
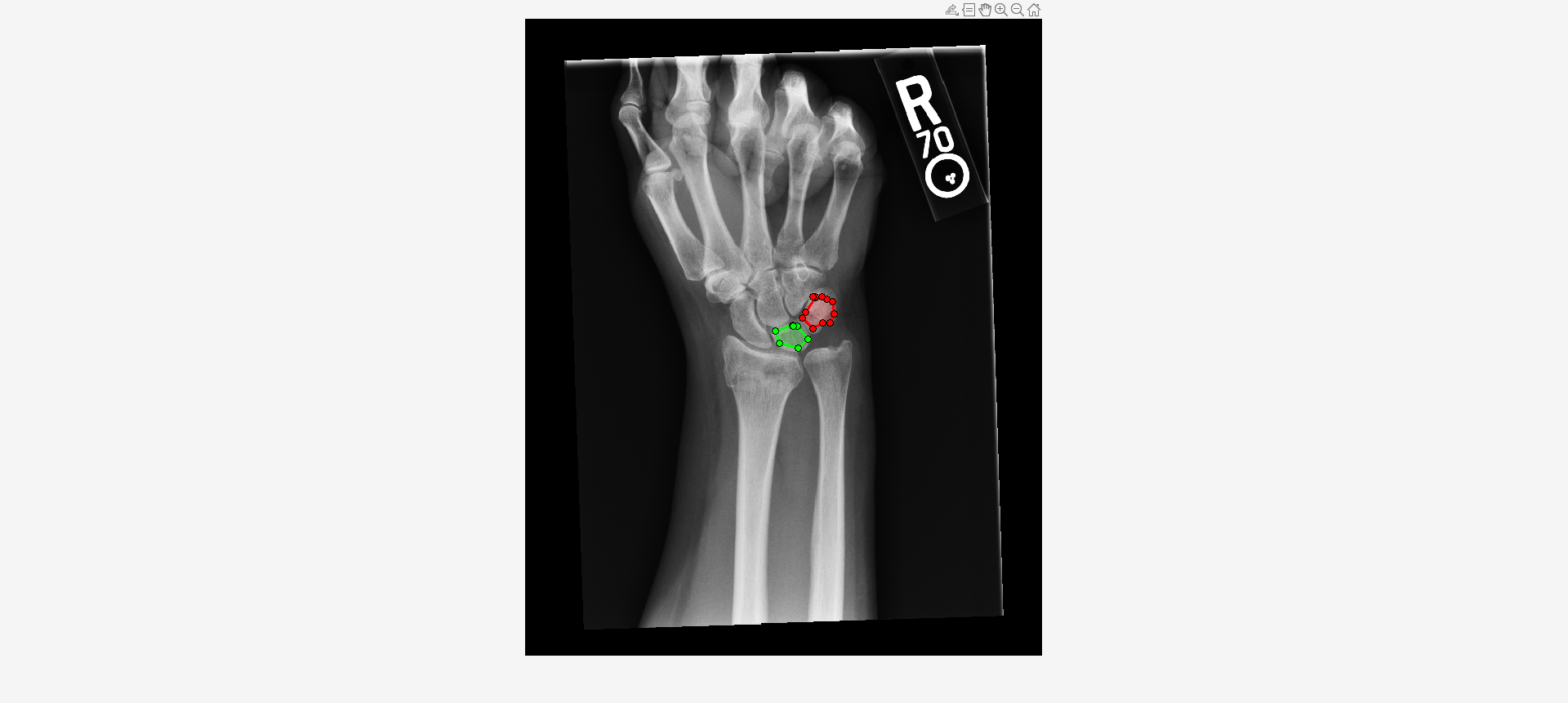
mask1 = createMask(roi1,I); mask2 = createMask(roi2,I);
Create an ROI label matrix, using different labels for the two ROIs. Create a medicalImage object of the ROI label data.
mask = zeros(size(I));
mask(mask1) = 1;
mask(mask2) = 2;
info = dicominfo("forearmXrayImage1.dcm");
roi = medicalImage(mask,info);Create a radiomics object from the X-ray image data and ROI label data.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalImage]
ROILabel: [1×1 medicalImage]
Resample: 0
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: []
DataResampleMethod: []
MaskResampleMethod: []
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute selected features for both ROIs.
F = selectFeatures(R,["MeanIntensity2D","ContrastAveraged2D"])
F=2×3 table
LabelID MeanIntensity2D ContrastAveraged2D
_______ _______________ __________________
"1" 2314.4 122.85
"2" 2438.5 104.57
Import a computed tomography (CT) image volume and the corresponding ROI mask volume from the IBSI validation data set [1][2][3] as medicalVolume objects.
unzip("CTImageMaskNIfTI.zip") data = medicalVolume("CT_image.nii.gz"); roi = medicalVolume("CT_mask.nii.gz");
Visualize a slice of the CT image volume and the corresponding ROI.
figure
imshowpair(data.Voxels(:,:,20),roi.Voxels(:,:,20),"montage")
Create a radiomics object, using the CT image volume and ROI mask volume, with default preprocessing options.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalVolume]
ROILabel: [1×1 medicalVolume]
Resample: 1
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: 1
DataResampleMethod: 'linear'
MaskResampleMethod: 'linear'
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute the selected features of the ROI in the CT image volume.
I = selectFeatures(R,["Sphericity3D","IntensityHistogramMode3D","Busyness25D"])
I=1×4 table
LabelID Sphericity3D IntensityHistogramMode3D Busyness25D
_______ ____________ ________________________ ___________
"1" 0.80911 10 10.072
[1] Vallières, Martin, Carolyn R. Freeman, Sonia R. Skamene, and Issam El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” The Cancer Imaging Archive, 2015. https://doi.org/10.7937/K9/TCIA.2015.7GO2GSKS.
[2] Vallières, M, C R Freeman, S R Skamene, and I El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” Physics in Medicine and Biology 60, no. 14 (July 7, 2015): 5471–96. https://doi.org/10.1088/0031-9155/60/14/5471.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository.” Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
Input Arguments
Names of the features to compute, specified as a string scalar, character vector, string array, or cell array of character vectors. To compute a single feature, specify its name as a string scalar or a character vector. To compute multiple features, specify their names as a string array or a cell array of character vectors. The names of the features must match the exact spellings, including the suffix representing subtype, as specified in the Corresponding Feature columns of the tables on the IBSI Standard and Radiomics Function Feature Correspondences page.
Data Types: char | string
Output Arguments
Selected features, returned as a table. The first column in F is
LabelID. The subsequent columns are the selected features. Each row
of the table corresponds to an ROI.
Version History
Introduced in R2025a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)