Medical Image Preprocessing
Image preprocessing prepares data for a target workflow. The main goals of medical image preprocessing are to reduce image acquisition artifacts and to standardize images across a data set. Your exact preprocessing requirements depend on the modality and procedure used to acquire data, as well as your target workflow. Some common preprocessing steps include background removal, denoising, resampling, registration, and intensity normalization.
Background Removal
Background removal involves segmenting the region of interest from the image background. By limiting the image to the region of interest, you can improve the efficiency and accuracy of your target workflow. One example of background removal is skull stripping, which removes the skull and other background regions from MRI images of the brain. Background removal typically consists of applying a mask of the region of interest that you create using morphological operations or other segmentation techniques. For more information about morphological operations, see Types of Morphological Operations.
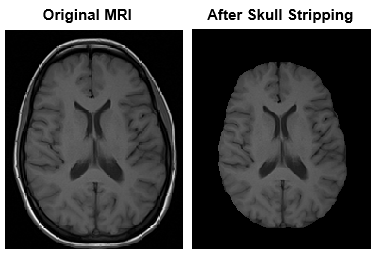
To perform background removal, multiply the mask image and the original image. For
example, consider a grayscale image, im, and a mask image,
mask, that is the same size as im and has a value of
1 for every element in the region of interest and 0
for each element of the background. This code returns a new image, imROI,
in which the elements in the region of interest have the same values as in
im, and the background elements all have values of
0.
imROI = im.*mask;
Denoising
Medical imaging modalities are susceptible to noise, which introduces random intensity
fluctuations in an image. To reduce noise, you can filter images in the spatial and
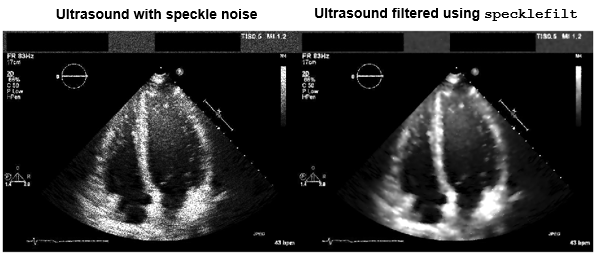
frequency domains. Medical Imaging Toolbox™ provides the specklefilt
function, which reduces the speckle noise common in ultrasound images. For additional image
filtering tools, see Image Filtering in Image Processing Toolbox™. You can also denoise medical image data using deep learning. For details, see
Train and Apply Denoising Neural Networks.
Resampling
Use resampling to change the pixel or voxel size of an image without changing its spatial limits in the patient coordinate system. Resampling is useful for standardizing image resolution across a data set that contains images from multiple scanners.
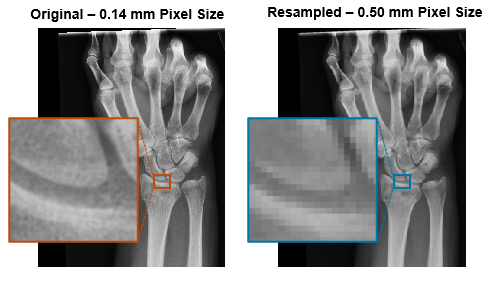
To resample 3-D image data in a medicalVolume
object, use the resample
object function. Using the resample object function maintains the
spatial referencing of the volume.
To resample a 2-D medical image, you can use the imresize
function with a target number of columns and rows. For example, this code shows how to
resample the pixel data in the medicalImage
object medImg to a target pixel size, specified by
targetPixelSpacing.
ratios = medImg.PixelSpacing./targetPixelSpacing;
targetSize = ceil(ratios.*size(medImg.Pixels)); % in pixels
resamplePixels = imresize(medImg.Pixels,targetSize);Define the spatial referencing of the original and resampled images by using the imref2d object.
originalR = imref2d(size(medImg.Pixels),medImg.PixelSpacing(1),medImg.PixelSpacing(2)); resampledR = imref2d(size(resamplePixels),targetPixelSpacing(1),targetPixelSpacing(2));
Registration
You can use image registration to standardize the spatial alignment of 2-D or 3-D medical images in a data set. Registration can be useful for aligning images of different patients, or images of the same patient acquired at different times, on different scanners, or using different imaging modalities. For more details, see Medical Image Registration.

Intensity Normalization
Intensity normalization standardizes the range of image intensity values across a data
set. Typically, you perform this process in two steps. First, clip intensities to a smaller
range. Second, normalize the clipped intensity range to the range of the image data type,
such as [0, 1] for double or [0, 255] for uint8.
Whereas visualizing image data using a display window changes how you view the data,
intensity normalization actually updates the image values.
One normalization strategy is to rescale the intensity values within each image relative
to the minimum and maximum values in the image. For example, this code uses the rescale
function to limit the intensities in the image im to the 0.5 and 99.5
percentile values, and to rescale the values to the range [0, 1]:
imMax = prctile(im(:),99.5); imMin = prctile(im(:),0.5); imNormalized = rescale(im,0,1,InputMin=imMin,InputMax=imMax);
Another strategy is to rescale values to the same intensity range across images. In CT
imaging, intensity windowing limits intensity values, in Hounsfield
units (HU), to a suitable range for the tissue of interest. HU is a standard CT density
scale defined by the density of air (–1000 HU) and water (0 HU). The
medicalVolume object automatically rescales the data returned in the
Voxels property for DICOM files that define the
RescaleIntercept and RescaleSlope metadata
attributes. You can find suggested intensity windows for your application in clinical
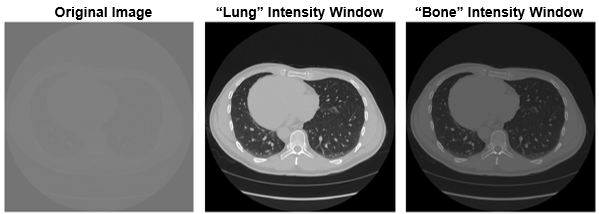
guidelines or from your scanner manufacturer. For example, to segment lungs from a chest CT,
you might use the range [–1200, 600] HU, and to segment bone, which is more dense, you might
use the range [–1400, 2400]. This figure shows a transverse slice of a chest CT with no
intensity windowing, a "lung" intensity window, and a "bone" intensity window.
This code applies intensity windowing to the image stored in the DICOM file
dicomFile, assuming the values of medVol.Voxels are
in units of
HU:
medVol = medicalVolume(dicomFile); lungWindow = [-1200 600]; imWindowed = rescale(medVol.Voxels,0,1,InputMin=lungWindow(1),InputMax=lungWindow(2));
Preprocessing in Advanced Workflows
For an example that preprocesses image data as part of a deep learning workflow, see Brain MRI Segmentation Using Pretrained 3-D U-Net Network.
See Also
specklefilt | resample | rescale | medicalVolume