sbiopredictionci
Compute confidence intervals for model predictions (requires Statistics and Machine Learning Toolbox)
Description
ci = sbiopredictionci(fitResults)fitResults, an NLINResults or OptimResults returned by sbiofit. ci is
a PredictionConfidenceInterval object that contains the computed
confidence interval data.
ci = sbiopredictionci(fitResults,Name,Value)Name,Value
pair arguments.
Examples
Load Data
Load the sample data to fit. The data is stored as a table with variables ID , Time , CentralConc , and PeripheralConc. This synthetic data represents the time course of plasma concentrations measured at eight different time points for both central and peripheral compartments after an infusion dose for three individuals.
load data10_32R.mat gData = groupedData(data); gData.Properties.VariableUnits = {'','hour','milligram/liter','milligram/liter'}; sbiotrellis(gData,'ID','Time',{'CentralConc','PeripheralConc'},'Marker','+',... 'LineStyle','none');

Create Model
Create a two-compartment model.
pkmd = PKModelDesign; pkc1 = addCompartment(pkmd,'Central'); pkc1.DosingType = 'Infusion'; pkc1.EliminationType = 'linear-clearance'; pkc1.HasResponseVariable = true; pkc2 = addCompartment(pkmd,'Peripheral'); model = construct(pkmd); configset = getconfigset(model); configset.CompileOptions.UnitConversion = true;
Define Dosing
Define the infusion dose.
dose = sbiodose('dose','TargetName','Drug_Central'); dose.StartTime = 0; dose.Amount = 100; dose.Rate = 50; dose.AmountUnits = 'milligram'; dose.TimeUnits = 'hour'; dose.RateUnits = 'milligram/hour';
Define Parameters
Define the parameters to estimate. Set the parameter bounds for each parameter. In addition to these explicit bounds, the parameter transformations (such as log, logit, or probit) impose implicit bounds.
responseMap = {'Drug_Central = CentralConc','Drug_Peripheral = PeripheralConc'};
paramsToEstimate = {'log(Central)','log(Peripheral)','Q12','Cl_Central'};
estimatedParam = estimatedInfo(paramsToEstimate,...
'InitialValue',[1 1 1 1],...
'Bounds',[0.1 3;0.1 10;0 10;0.1 2]);
Fit Model
Perform an unpooled fit, that is, one set of estimated parameters for each patient.
unpooledFit = sbiofit(model,gData,responseMap,estimatedParam,dose,'Pooled',false);
Perform a pooled fit, that is, one set of estimated parameters for all patients.
pooledFit = sbiofit(model,gData,responseMap,estimatedParam,dose,'Pooled',true);
Compute Confidence Intervals for Estimated Parameters
Compute 95% confidence intervals for each estimated parameter in the unpooled fit.
ciParamUnpooled = sbioparameterci(unpooledFit);
Display Results
Display the confidence intervals in a table format. For details about the meaning of each estimation status, see Parameter Confidence Interval Estimation Status.
ci2table(ciParamUnpooled)
ans =
12×7 table
Group Name Estimate ConfidenceInterval Type Alpha Status
_____ ______________ ________ __________________ ________ _____ ___________
1 {'Central' } 1.422 1.1533 1.6906 Gaussian 0.05 estimable
1 {'Peripheral'} 1.5629 0.83143 2.3551 Gaussian 0.05 constrained
1 {'Q12' } 0.47159 0.20093 0.80247 Gaussian 0.05 constrained
1 {'Cl_Central'} 0.52898 0.44842 0.60955 Gaussian 0.05 estimable
2 {'Central' } 1.8322 1.7893 1.8751 Gaussian 0.05 success
2 {'Peripheral'} 5.3368 3.9133 6.7602 Gaussian 0.05 success
2 {'Q12' } 0.27641 0.2093 0.34351 Gaussian 0.05 success
2 {'Cl_Central'} 0.86034 0.80313 0.91755 Gaussian 0.05 success
3 {'Central' } 1.6657 1.5818 1.7497 Gaussian 0.05 success
3 {'Peripheral'} 5.5632 4.7557 6.3708 Gaussian 0.05 success
3 {'Q12' } 0.78361 0.65581 0.91142 Gaussian 0.05 success
3 {'Cl_Central'} 1.0233 0.96375 1.0828 Gaussian 0.05 success
Plot the confidence intervals. If the estimation status of a confidence interval is success, it is plotted in blue (the first default color). Otherwise, it is plotted in red (the second default color), which indicates that further investigation into the fitted parameters may be required. If the confidence interval is not estimable, then the function plots a red line with a centered cross. If there are any transformed parameters with estimated values 0 (for the log transform) and 1 or 0 (for the probit or logit transform), then no confidence intervals are plotted for those parameter estimates. To see the color order, type get(groot,'defaultAxesColorOrder').
Groups are displayed from left to right in the same order that they appear in the GroupNames property of the object, which is used to label the x-axis. The y-labels are the transformed parameter names.
plot(ciParamUnpooled)

Compute the confidence intervals for the pooled fit.
ciParamPooled = sbioparameterci(pooledFit);
Display the confidence intervals.
ci2table(ciParamPooled)
ans =
4×7 table
Group Name Estimate ConfidenceInterval Type Alpha Status
______ ______________ ________ __________________ ________ _____ ___________
pooled {'Central' } 1.6626 1.3287 1.9965 Gaussian 0.05 estimable
pooled {'Peripheral'} 2.687 0.89848 4.8323 Gaussian 0.05 constrained
pooled {'Q12' } 0.44956 0.11445 0.85152 Gaussian 0.05 constrained
pooled {'Cl_Central'} 0.78493 0.59222 0.97764 Gaussian 0.05 estimable
Plot the confidence intervals. The group name is labeled as "pooled" to indicate such fit.
plot(ciParamPooled)

Plot all the confidence interval results together. By default, the confidence interval for each parameter estimate is plotted on a separate axes. Vertical lines group confidence intervals of parameter estimates that were computed in a common fit.
ciAll = [ciParamUnpooled;ciParamPooled]; plot(ciAll)
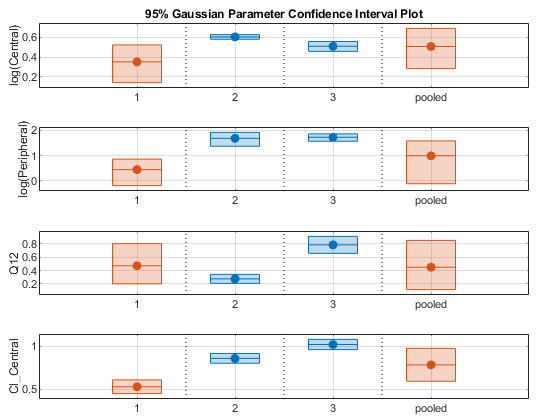
You can also plot all confidence intervals in one axes grouped by parameter estimates using the 'Grouped' layout.
plot(ciAll,'Layout','Grouped')
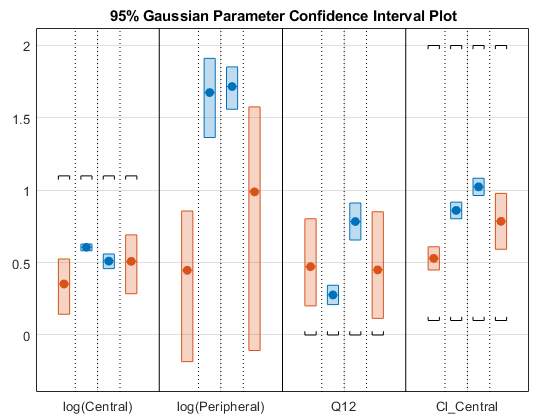
In this layout, you can point to the center marker of each confidence interval to see the group name. Each estimated parameter is separated by a vertical black line. Vertical dotted lines group confidence intervals of parameter estimates that were computed in a common fit. Parameter bounds defined in the original fit are marked by square brackets. Note the different scales on the y-axis due to parameter transformations. For instance, the y-axis of Q12 is in the linear scale, but that of Central is in the log scale due to its log transform.
Compute Confidence Intervals for Model Predictions
Calculate 95% confidence intervals for the model predictions, that is, simulation results using the estimated parameters.
% For the pooled fit ciPredPooled = sbiopredictionci(pooledFit); % For the unpooled fit ciPredUnpooled = sbiopredictionci(unpooledFit);
Plot Confidence Intervals for Model Predictions
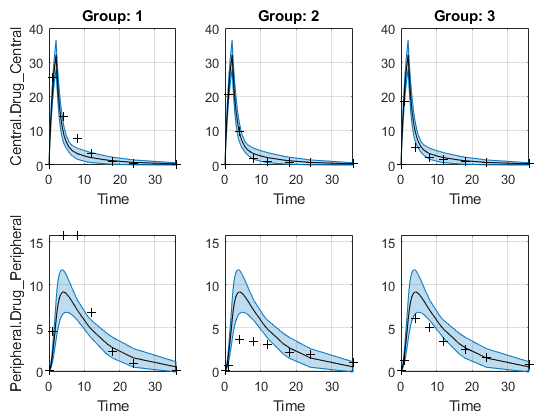
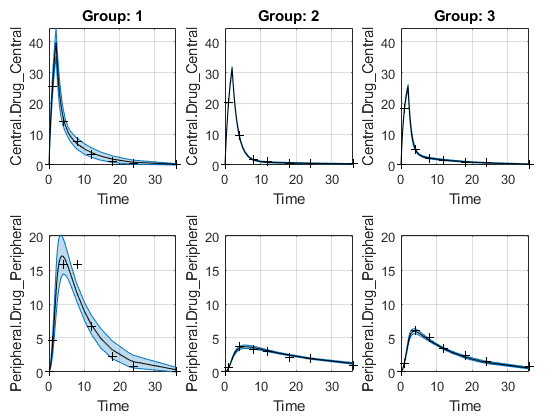
The confidence interval for each group is plotted in a separate column, and each response is plotted in a separate row. Confidence intervals limited by the bounds are plotted in red. Confidence intervals not limited by the bounds are plotted in blue.
plot(ciPredPooled)
plot(ciPredUnpooled)
Input Arguments
Parameter estimation results from sbiofit, specified as an NLINResults, OptimResults, or a vector of objects for unpooled fits
that were returned from the same sbiofit call.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: 'Alpha',0.01,'Type','bootstrap' specifies to compute a
99% confidence interval using the bootstrap method.
Confidence level, (1-Alpha) * 100%,
specified as the comma-separated pair consisting of 'Alpha' and a
positive scalar between 0 and 1. The default value is 0.05,
meaning a 95% confidence interval is computed.
Example: 'Alpha',0.01
Confidence interval type, specified as the comma-separated pair consisting of
'Type' and a character vector. The valid choices are:
'gaussian'– Use the Gaussian approximation of the distribution of the linearized model responses around the parameter estimates.'bootstrap'– Compute confidence intervals using the bootstrap method.
Example: 'Type','bootstrap'
Number of samples for bootstrapping, specified as the comma-separated pair
consisting of 'NumSamples' and a positive integer. This number
defines the number of fits that are performed during the confidence interval
computation to generate bootstrap samples. The smaller the number is, the faster the
computation of the confidence intervals becomes, at the cost of decreased
accuracy.
Example: 'NumSamples',500
Level of display returned to the command line, specified as the comma-separated pair
consisting of 'Display' and a character vector.
'off' (default) or 'none' displays no
output. 'final' displays a message when the computation
finishes.
Example: 'Display','final'
Logical flag to compute confidence intervals in parallel, specified as the
comma-separated pair consisting of 'UseParallel' and
true or false. By default, the parallel
options in the original fit are used. If this argument is set to
true and Parallel Computing Toolbox™ is available, the parallel options in the original fit are ignored,
and confidence intervals are computed in parallel.
For the Gaussian confidence intervals:
If the input
fitResultsis a vector of results objects, then the computation of confidence intervals for each object is performed in parallel. The Gaussian confidence intervals are quick to compute. So, it might be more beneficial to parallelize the original fit (sbiofit) and not setUseParallelto true forsbiopredictionci.
For the Bootstrap confidence intervals:
The function forwards the
UseParallelflag tobootci. There is no parallelization over the input vector of results objects.
Note
If you have a global stream for random number generation with a number of
substreams to compute in parallel in a reproducible fashion,
sbiopredictionci first checks to see if the number
of workers is same as the number of substreams. If so, the function sets
UseSubstreams to true in the
statset option and passes to bootci (Statistics and Machine Learning Toolbox). Otherwise, the
substreams are ignored by default.
Example: 'UseParallel',true
Output Arguments
Confidence interval results, returned as a PredictionConfidenceInterval object. For an unpooled fit,
ci can be a vector of
PredictionConfidenceInterval objects.
More About
The model is linearized around the parameter estimates
Pest that are obtained from the
fit results returned by sbiofit. The
CovarianceMatrix is transformed using the linearized model.
In addition, implicit parameter bounds (log,
probit, or logit parameter transforms
specified in the original fit) and explicit parameter bounds (if specified in the
original fit) are also mapped through the linearized model.
To linearize the model, sbiopredictionci first checks to see
if the sensitivity analysis feature is turned on in the original fit. If the feature
is on, the function uses the Jacobian computed via the complex step differentiation.
If the feature is off, the Jacobian is computed using finite differencing. Finite
differencing can be inaccurate, and consider turning on the sensitivity
analysis feature when you run sbiofit.
The function uses the transformed CovarianceMatrix and
computes the Gaussian confidence intervals for each estimated model response at
every time step.
In cases where the confidence interval is constrained by the parameter bounds defined in the original fit, the confidence interval bounds are adjusted according to the approach described by Wu, H. and Neale, M. [1].
For each model response, the function first decides whether the confidence interval is unbounded. If so, the estimation status of the corresponding model response is set to
not estimable.Otherwise, if the confidence interval for a response is constrained by a parameter bound defined in the original fit, the function sets its status to
constrained. Parameter transformations (such aslog,probit, orlogit) impose implicit bounds on the estimated parameters, for example, positivity constraints. Such bounds can lead to the overestimation of confidence, that is, the confidence interval can be smaller than expected.If no confidence interval has the estimation status
not estimableorconstrained, then the function sets the estimation statuses of all model responses tosuccess. Otherwise, the estimation statuses of remaining model responses are set toestimable.
The bootci (Statistics and Machine Learning Toolbox) function from Statistics and Machine Learning Toolbox™ is used to compute the bootstrap confidence intervals. The first input
nboot is the number of samples
(NumSamples), and the second input bootfun is
a function that performs these actions.
Resample the data (independently within each group, if multiple groups are available).
Run a parameter fit with the resampled data.
Simulate the model using the estimated parameters to get model responses.
Return model responses.
The estimation status is always set to estimable since the
function cannot determine if the confidence intervals are constrained by the
bounds on the parameter estimates.
References
[1] Wu, H., and M.C. Neale. "Adjusted Confidence Intervals for a Bounded Parameter." Behavior Genetics. 42 (6), 2012, pp. 886-898.
Extended Capabilities
To run in parallel, set 'UseParallel' to true.
For more information, see the 'UseParallel' name-value pair argument.
Version History
Introduced in R2017b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)