cophenet
Cophenetic correlation coefficient
Description
Examples
Load the examgrades data set.
load examgradesCreate a hierarchical cluster tree using the linkage function. Specify the average method and the Minkowski distance metric with an exponent of 3.
Z = linkage(grades,"average",{"minkowski",3});
Compute the distances between pairs of observations using the pdist function.
Y = pdist(grades);
Compute the cophenetic correlation coefficient.
c = cophenet(Z,Y)
c = 0.6308
The moderately high correlation coefficient suggests that the hierarchical clustering tree provides a reasonably good representation of the distances between observations.
Create a sample data set consisting of randomly generated data from three standard uniform distributions.
rng(0,"twister"); % For reproducibility X = [gallery("uniformdata",[10 3],12); ... gallery("uniformdata",[10 3],13)+1.2; ... gallery("uniformdata",[10 3],14)+2.5]; c = [ones(10,1);2*(ones(10,1));3*(ones(10,1))]; % Actual classes
Create a scatter plot of the data.
scatter3(X(:,1),X(:,2),X(:,3),100,c,"filled")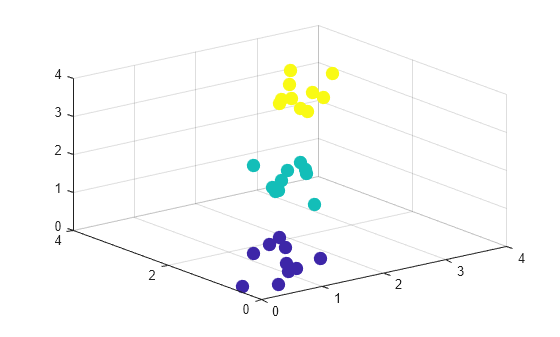
Create a hierarchical cluster tree using the linkage function. Specify the weighted method and the standardized Euclidean distance metric.
Z = linkage(X,"weighted","seuclidean");
Compute the distances between pairs of observations using the pdist function, and display a dendrogram plot.
Y = pdist(X); dendrogram(Z)
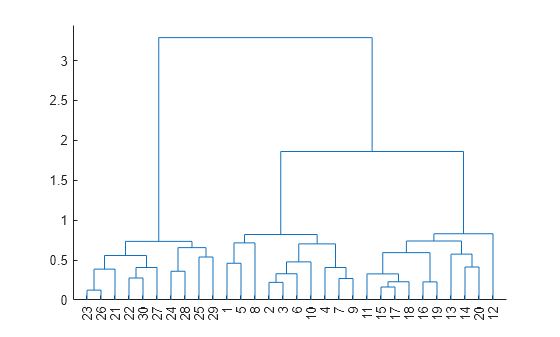
Return the cophenetic correlation coefficient and cophenetic distances.
[c,D] = cophenet(Z,Y)
c = 0.8179
D = 1×435
0.8203 0.8203 0.8203 0.4604 0.8203 0.8203 0.7150 0.8203 0.8203 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 3.2866 3.2866 3.2866 3.2866 3.2866 3.2866 3.2866 3.2866 3.2866 3.2866 0.2213 0.7024 0.8203 0.3286 0.7024 0.8203 0.7024 0.4772 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 1.8599 3.2866 3.2866 3.2866
The high correlation coefficient suggests that the dendrogram provides a good representation of the pairwise distances Y.
Create a second hierarchical cluster tree using the complete method, which computes the largest distance between objects in each cluster.
ZZ = linkage(X,"complete","seuclidean");
Compute the distances between pairs of observations. Return the cophenetic correlation coefficient and the cophenetic distances.
YY = pdist(X); [cc,DD] = cophenet(ZZ,YY)
cc = 0.8202
DD = 1×435
1.2044 1.2044 1.2044 0.4604 1.2044 1.2044 1.2044 1.2044 1.2044 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 5.0417 5.0417 5.0417 5.0417 5.0417 5.0417 5.0417 5.0417 5.0417 5.0417 0.2213 0.8986 1.2044 0.3696 0.8986 0.8595 0.8986 0.5287 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 2.9605 5.0417 5.0417 5.0417
Create a scatter plot of pairwise distance versus cophenetic distance for the two cluster trees.
scatter(D,Y) hold on scatter(DD,YY,"x") plot([0,max(Y)],[0,max(Y)],"b:",LineWidth=2); % Plot the 1:1 line xlabel("Cophenetic Distance"); ylabel("Pairwise Distance") legend("Weighted","Complete","1:1 line",Location="northwest") hold off
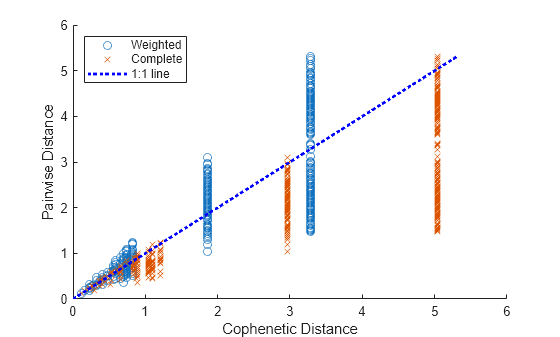
The cluster trees have similar cophenetic correlation coefficients, but the cophenetic distances of the tree created with the complete method are systematically larger than their corresponding pairwise distances.
Input Arguments
Hierarchical cluster tree, specified as a numeric matrix returned by the linkage function. Z has size (m –
1)-by-3, where m is the number of observations used to create the
cluster tree. The third column of Z contains linkage distances. For
more information, see Agglomerative
hierarchical cluster tree.
Data Types: single | double
Distances (or dissimilarities) used to create Z, specified as a
numeric row vector returned by the pdist function. Y has length
m*(m – 1)/2, where m is the
number of observations used to create the cluster tree.
Data Types: single | double
Output Arguments
Cophenetic correlation coefficient, returned as a numeric scalar. The cophenetic correlation for a cluster tree is the linear correlation coefficient between the cophenetic distances obtained from the tree, and the original distances (or dissimilarities) used to create the tree. So, the cophenetic correlation coefficient is a measure of how faithfully the tree represents the dissimilarities among observations. A cophenetic correlation coefficient with a magnitude close to 1 indicates a high-quality solution. You can use this measure to compare alternative cluster solutions obtained using different algorithms.
The cophenetic correlation between Z(:,3) and
Y is defined as
where:
Yij is the distance between objects i and j in
Y.Zij is the cophenetic distance between objects i and j, from
Z(:,3).y and z are the averages of
YandZ(:,3), respectively.
Cophenetic distances, returned as a numeric row vector with the same length as
Y. The cophenetic distance between two observations is
represented in a dendrogram by the height of the link at which the two observations are
first joined. This height is the distance between the two subclusters that are merged by
the link.
Version History
Introduced before R2006a
See Also
cluster | dendrogram | inconsistent | linkage | pdist | squareform
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)