cluster
Construct agglomerative clusters from linkages
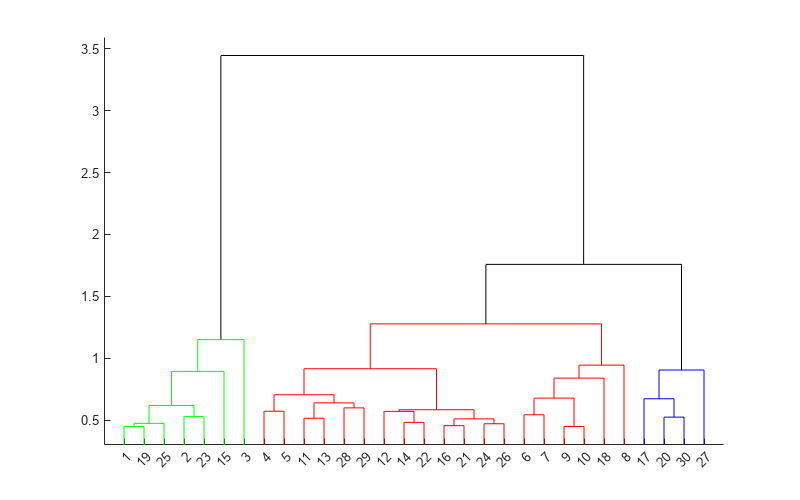
Description
T = cluster(Z,Cutoff=cutoff)Z.
The input Z is the output of the linkage function for an input data matrix X.
cluster cuts Z into clusters, using
cutoff as a threshold for the inconsistency coefficients (or
inconsistent values) of nodes in the tree. The
output T contains cluster assignments of each observation (row of
X).
T = cluster(___,Name=Value)cluster(Z,MaxClust=5,Depth=3) to find a maximum of five clusters by
evaluating distance values up to a depth of three below each node.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Alternative Functionality
If you have an input data matrix X, you can use clusterdata to perform agglomerative clustering and return cluster indices for
each observation (row) in X. The clusterdata function
performs all the necessary steps for you, so you do not need to execute the pdist, linkage, and cluster
functions separately.