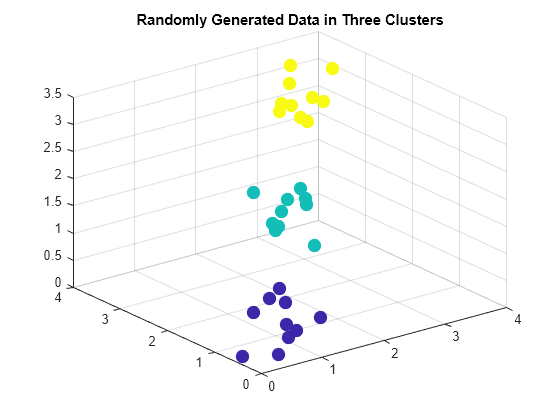
clusterdata
Construct agglomerative clusters from data
Syntax
Description
T = clusterdata(X,Cutoff=cutoff)X, given a threshold cutoff for cutting an
agglomerative hierarchical tree generated by the linkage function from X.
clusterdata supports agglomerative clustering and incorporates
the pdist, linkage, and
cluster functions, which you can use
separately for more detailed analysis. See Algorithm Description for more details.
T = clusterdata(___,Name=Value)clusterdata(X,MaxClust=5,Depth=3) to find a maximum of five clusters
by evaluating distance values up to a depth of three below each node.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Tips
If
Linkageis"centroid"or"median", thenlinkagecan produce a cluster tree that is not monotonic. This result occurs when the distance from the union of two clusters, r and s, to a third cluster is less than the distance between r and s. In this case, in a dendrogram drawn with the default orientation, the path from a leaf to the root node takes some downward steps. To avoid this result, specify another value forLinkage. The following image shows a nonmonotonic cluster tree.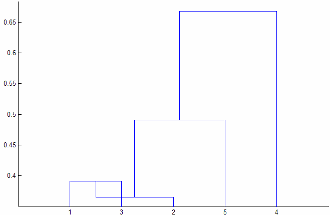
In this case, cluster 1 and cluster 3 are joined into a new cluster, while the distance between this new cluster and cluster 2 is less than the distance between cluster 1 and cluster 3.
Algorithms
When you do not specify any optional name-value arguments, the
clusterdata function performs the following steps:
Create a vector of the Euclidean distance between pairs of observations in
Xby usingpdist.Y =pdist(X,"euclidean")Create an agglomerative hierarchical cluster tree from
Yby usinglinkagewith the"single"method for computing the shortest distance between clusters.Z =linkage(Y,"single")When you specify
cutoff, theclusterdatafunction usesclusterto define clusters fromZwhen inconsistent values are less thancutoff.T=cluster(Z,Cutoff=cutoff)When you specify
maxclust, theclusterdatafunction usesclusterto find a maximum ofmaxclustclusters fromZ, using"distance"as the criterion for defining clusters.T= cluster(Z,MaxClust=maxclust)
Alternative Functionality
If you have a hierarchical cluster tree Z (the output of the linkage function for the input data matrix X), you can use
cluster to perform agglomerative clustering on Z and return
the cluster assignment for each observation (row) in X.