fit
Train robust random cut forest model for incremental anomaly detection
Since R2023b
Syntax
Description
The fit function fits a configured robust random cut forest
(RRCF) model for incremental anomaly detection (incrementalRobustRandomCutForest object) to streaming data.
To fit a RRCF model to an entire batch of data at once, see rrcforest.
forest = fit(forest,Tbl)forest, which represents the
input incremental learning model forest trained using the predictor
data in Tbl. Specifically, the fit
function fits the model to the incoming data and stores the updated score threshold and
configurations in the output model forest.
Examples
Prepare an incremental robust random cut forest model by specifying an anomaly contamination fraction of 0.001, and standardize the data using an initial estimation period of 500 observations. Specify a score warm-up period of 1000 observations, during which the fit function updates the score threshold and trains the model but does not return scores or identify anomalies.
forest = incrementalRobustRandomCutForest(ContaminationFraction=0.001, ...
StandardizeData=true,ScoreWarmupPeriod=1000,EstimationPeriod=500);forest is an incrementalRobustRandomCutForest model object. All its properties are read-only. forest must be fit to data before you can use it to perform any other operations.
Load Data
Load the credit rating data stored in CreditRating_Historical.dat. Remove the ID column and the categorical variables.
creditrating = readtable("CreditRating_Historical.dat"); creditrating = removevars(creditrating,["ID","Industry","Rating"]);
The fit function of incrementalRobustRandomCutForest does not use observations with missing values. Remove missing values in the data sets to reduce memory consumption and speed up training.
creditrating = rmmissing(creditrating);
Fit Incremental Model and Detect Anomalies
Fit the incremental model to the data by using the fit function. To simulate a data stream, fit the model in chunks of 100 observations at a time. Because EstimationPeriod = 500 and ScoreWarmupPeriod = 1000, fit only returns scores and detects anomalies after 15 iterations. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
meanscore, the mean score value of the data chunk, to see how it evolves during incremental learning.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the chunk, to see how it evolves during incremental learning.
n = numel(creditrating(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); meanscore = zeros(nchunk,1); threshold = zeros(nchunk,1); numAnom = zeros(nchunk,1); % Incremental fitting rng(0,"twister"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; [forest,tf,scores] = fit(forest,creditrating(idx,:)); meanscore(j) = mean(scores); numAnom(j) = sum(tf); threshold(j) = forest.ScoreThreshold; end
forest is an incrementalRobustRandomCutForest model object trained on all the data in the stream.
Analyze Incremental Model During Training
To see how the mean score, score threshold and number of detected anomalies per chunk evolve during training, plot them on separate tiles.
tiledlayout(3,1); nexttile plot(meanscore) ylabel("Mean Score") xlabel("Iteration") xlim([0 nchunk]) xline(forest.EstimationPeriod/numObsPerChunk,"r-.") xline((forest.EstimationPeriod+forest.ScoreWarmupPeriod)/numObsPerChunk,"r") nexttile plot(threshold) ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk]) xline(forest.EstimationPeriod/numObsPerChunk,"r-.") xline((forest.EstimationPeriod+forest.ScoreWarmupPeriod)/numObsPerChunk,"r") nexttile plot(numAnom,"+") ylabel("Anomalies") xlabel("Iteration") xlim([0 nchunk]) ylim([0 max(numAnom)+0.2]) xline(forest.EstimationPeriod/numObsPerChunk,"r-.") xline((forest.EstimationPeriod+forest.ScoreWarmupPeriod)/numObsPerChunk,"r")
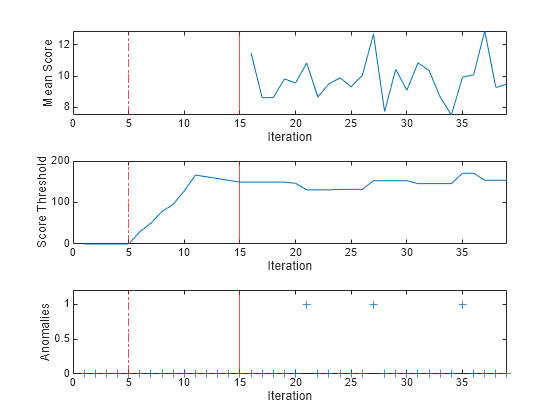
During the estimation period, fit estimates means and standard deviations using the observations, and does not fit the model or update the score threshold. During the warm-up period, fit fits the model and updates the score threshold, but returns all scores as NaN and all anomaly values as false. After the warm-up period, fit returns the observation scores and the indices of observations with scores above the score threshold value. A small score value indicates a normal observation, and a large score value indicates an anomaly.
totalAnomalies=sum(numAnom)
totalAnomalies = 3
anomfrac= totalAnomalies/(n-forest.EstimationPeriod-forest.ScoreWarmupPeriod)
anomfrac = 0.0012
The software detects 3 anomalies after the warm-up and estimation periods. The contamination fraction after the estimation and warm-up periods is approximately 0.001.
Train a robust random cut forest (RRCF) model on a simulated, noisy, periodic shingled time series containing no anomalies by using rrcforest. Convert the trained model to an incremental learner object, and then incrementally fit the time series and detect anomalies.
Create Simulated Data Stream
Create a simulated data stream of observations representing a noisy sinusoid signal.
rng(0,"twister"); % For reproducibility period = 100; n = 2001+period; sigma = 0.04; a = linspace(1,n,n)'; b = sin(2*pi*(a-1)/period)+sigma*randn(n,1);
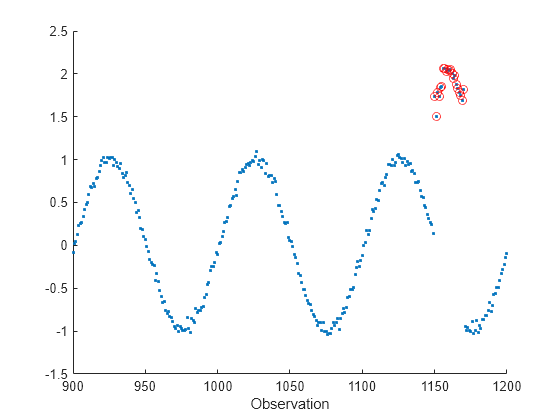
Introduce an anomalous region into the data stream. Plot the data stream portion that contains the anomalous region, and circle the anomalous data points.
c = 2*(sin(2*pi*(a-35)/period)+sigma*randn(n,1)); b(1150:1170) = c(1150:1170); scatter(a,b,".") xlim([900,1200]) xlabel("Observation") hold on scatter(a(1150:1170),b(1150:1170),"r") hold off
Convert the single-featured data set b into a multi-featured data set by shingling [1] with a shingle size equal to the period of the signal. The th shingled observation is a vector of features with values , , ..., , where is the shingle size.
X = []; shingleSize = period; for i = 1:n-shingleSize X = [X;b(i:i+shingleSize-1)']; end
Train Model and Perform Incremental Anomaly Detection
Fit a robust random cut forest model to the first 1000 shingled observations, specifying a contamination fraction of 0. Convert the model to an incrementalRobustRandomCutForest model object. Specify to keep the 100 most recent observations relevant for anomaly detection.
Mdl = rrcforest(X(1:1000,:),ContaminationFraction=0); IncrementalMdl = incrementalLearner(Mdl,NumObservationsToKeep=100);
To simulate a data stream, process the full shingled data set in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Calculate scores and detect anomalies using the
isanomalyfunction.Store
anomIdx, the indices of shingled observations marked as anomalies.If the chunk contains fewer than three anomalies, fit and update the previous incremental model.
n = numel(X(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); anomIdx = []; allscores = []; % Incremental fitting rng("default"); % For reproducibility for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; [isanom,scores] = isanomaly(IncrementalMdl,X(idx,:)); allscores = [allscores;scores]; anomIdx = [anomIdx;find(isanom)+ibegin-1]; if (sum(isanom) < 3) IncrementalMdl = fit(IncrementalMdl,X(idx,:)); end end
Analyze Incremental Model During Training
At each iteration, the software calculates a score value for each observation in the data chunk. A negative score value with large magnitude indicates a normal observation, and a large positive value indicates an anomaly. Plot the anomaly score for the observations in the vicinity of the anomaly. Circle the scores of shingles that the software returns as anomalous.
figure scatter(a(1:2000),allscores,".") hold on scatter(a(anomIdx),allscores(anomIdx),20,"or") xlim([900,1200]) xlabel("Shingle") ylabel("Score") hold off
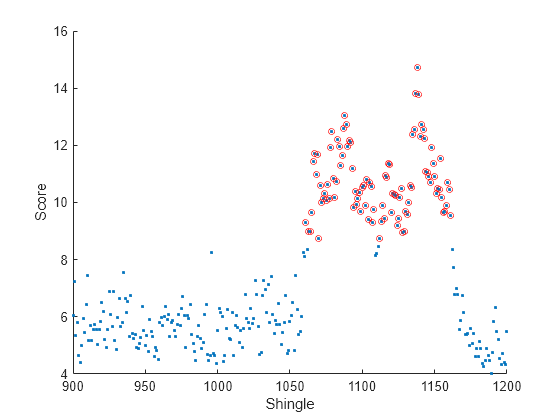
Because the introduced anomalous region begins at observation 1150, and the shingle size is 100, shingle 1051 is the first to show a high anomaly score. Some shingles between 1050 and 1170 have scores lying just below the anomaly score threshold, due to the noise in the sinusoidal signal. The shingle size affects the performance of the model by defining how many subsequent consecutive data points in the original time series the software uses to calculate the anomaly score for each shingle.
Plot the unshingled data and highlight the introduced anomalous region. Circle the observation number of the first element in each shingle returned by that the software as anomalous.
figure xlim([900,1200]) ylim([-1.5 2]) rectangle(Position=[1150 -1.5 20 3.5],FaceColor=[0.9 0.9 0.9], ... EdgeColor=[0.9 0.9 0.9]) hold on scatter(a,b,".") scatter(a(anomIdx),b(anomIdx),20,"or") xlabel("Observation") hold off

Train an incremental robust random cut forest (RRCF) model and perform anomaly detection on a data set with categorical predictors.
Load Data
Load census1994.mat. The data set consists of demographic data from the US Census Bureau.
load census1994.matincrementalRobustRandomCutForest does not use observations with missing values. Remove missing values in the data to reduce memory consumption and speed up training. Keep only the first 1000 observations in the training data set and the first 2000 observations in the test data set.
adultdata = rmmissing(adultdata); adulttest = rmmissing(adulttest); Xtrain = adultdata(1:1000,:); Xstream = adulttest(1:2000,:);
Train RRCF Model
Fit an RRCF model to the training data. Specify an anomaly contamination fraction of 0.001.
rng(0,"twister"); % For reproducibility TTforest = rrcforest(Xtrain,ContaminationFraction=0.001); details(TTforest)
RobustRandomCutForest with properties:
CollusiveDisplacement: 'maximal'
NumLearners: 100
NumObservationsPerLearner: 256
Mu: []
Sigma: []
CategoricalPredictors: [2 4 6 7 8 9 10 14 15]
ContaminationFraction: 1.0000e-03
ScoreThreshold: 55.5745
PredictorNames: {'age' 'workClass' 'fnlwgt' 'education' 'education_num' 'marital_status' 'occupation' 'relationship' 'race' 'sex' 'capital_gain' 'capital_loss' 'hours_per_week' 'native_country' 'salary'}
Methods, Superclasses
TTforest is a RobustRandomCutForest model object representing a traditionally trained RRCF model. The software identifies nine variables in the data as categorical predictors because they contain string arrays.
Convert Trained Model
Convert the traditionally trained RRCF model to an RRCF model for incremental learning.
Incrementalforest = incrementalLearner(TTforest);
Incrementalforest is an incrementalRobustRandomCutForest model object that is ready for incremental learning and anomaly detection.
Fit Incremental Model and Detect Anomalies
Perform incremental learning on the Xstream data by using the fit function. To simulate a data stream, fit the model in chunks of 100 observations at a time. At each iteration:
Process 100 observations.
Overwrite the previous incremental model with a new one fitted to the incoming observations.
Store
medianscore, the median score value of the data chunk, to see how it evolves during incremental learning.Store
threshold, the score threshold value for anomalies, to see how it evolves during incremental learning.Store
numAnom, the number of detected anomalies in the chunk, to see how it evolves during incremental learning.
n = numel(Xstream(:,1)); numObsPerChunk = 100; nchunk = floor(n/numObsPerChunk); medianscore = zeros(nchunk,1); numAnom = zeros(nchunk,1); threshold = zeros(nchunk,1); % Incremental fitting for j = 1:nchunk ibegin = min(n,numObsPerChunk*(j-1) + 1); iend = min(n,numObsPerChunk*j); idx = ibegin:iend; [Incrementalforest,tf,scores] = fit(Incrementalforest,Xstream(idx,:)); medianscore(j) = median(scores); numAnom(j) = sum(tf); threshold(j) = Incrementalforest.ScoreThreshold; end
Analyze Incremental Model During Training
To see how the median score, score threshold, and number of detected anomalies per chunk evolve during training, plot them on separate tiles.
tiledlayout(3,1); nexttile plot(medianscore) ylabel("Median Score") xlabel("Iteration") xlim([0 nchunk]) nexttile plot(threshold) ylabel("Score Threshold") xlabel("Iteration") xlim([0 nchunk]) nexttile plot(numAnom,"+") ylabel("Anomalies") xlabel("Iteration") xlim([0 nchunk]) ylim([0 max(numAnom)+0.2])

totalanomalies=sum(numAnom)
totalanomalies = 1
anomfrac= totalanomalies/n
anomfrac = 5.0000e-04
fit updates the model and returns the observation scores and the indices of observations with scores above the score threshold value as anomalies. A high score value indicates a normal observation, and a low value indicates an anomaly. The median score fluctuates between approximately 230 and 270. The score threshold rises from a value of 260 after the first iteration and steadily approaches 285 after 12 iterations. The software detected 4 anomalies in the Xstream data, yielding a total contamination fraction of 0.002.
Input Arguments
Incremental anomaly detection model to fit to streaming data, specified as an
incrementalRobustRandomCutForest model object. You can create
forest by calling
incrementalRobustRandomCutForest directly, or by
converting a supported, traditionally trained RRCF model using the incrementalLearner function.
Predictor data, specified as a table. Each row of Tbl
corresponds to one observation, and each column corresponds to one predictor
variable. Multicolumn variables and cell arrays other than cell arrays of
character vectors are not allowed.
If you train forest using a table, then you must provide
predictor data by using Tbl, not X. All
predictor variables in Tbl must have the same variable names
and data types as those in the training data. However, the column order in
Tbl does not need to correspond to the column order of the
training data.
Note
If an observation contains missing values for all predictors
(NaN, '' (empty character vector),
"" (empty string), <missing>, or
<undefined>) ,
fit
ignores the observation. Consequently,
fit
uses fewer than n observations to create an
updated model, where n is the number of observations in
Tbl.
Data Types: table
Predictor data, specified as a numeric matrix. Each row of
X corresponds to one observation, and each column
corresponds to one predictor variable.
If you train forest using a matrix, then you must provide
predictor data by using X, not Tbl. The
variables that make up the columns of X must have the same
order as the variables in the training data. If
forest.NumPredictors is not specified, then
fit infers it from the data.
Note
If an observation contains missing values for all predictors
(NaN) value, fit ignores the
observation. Consequently, fit uses fewer than
n observations to create an updated model, where
n is the number of observations in
X.
Data Types: single | double
Output Arguments
Updated RRCF model for incremental anomaly detection, returned as an incrementalRobustRandomCutForest model object.
Anomaly indicators, returned as a logical column vector. An element of
tf is true when the observation in the
corresponding row of Tbl or X is an
anomaly, and false otherwise. tf has the
same length as Tbl or X.
fit
updates forest and then identifies the observations
with scores above the threshold (the
ScoreThreshold value) as anomalies.
Note
If the model is not warm (
IsWarm=false), thenfitreturns alltfasfalse.fitassigns the anomaly indicator offalse(logical 0) to observations that have missing values for all predictors.
Data Types: logical
Anomaly scores, returned as a numeric column vector whose values are in the
range [0,Inf). scores has the same length as
Tbl or X, and each element of
scores contains an anomaly score for the observation in
the corresponding row of Tbl or X.
fit calculates scores after updating forest. A small
positive value indicates a normal observation, and a large positive value
indicates an anomaly.
Note
If the model is not warm (
IsWarm=false), thenfitreturns allscoresasNaN.fitassigns the anomaly score ofNaNto observations that have missing values for all predictors.
Data Types: single | double
References
[1] Guha, Sudipto, N. Mishra, G. Roy, and O. Schrijvers. "Robust Random Cut Forest Based Anomaly Detection on Streams," Proceedings of The 33rd International Conference on Machine Learning 48 (June 2016): 2712–21.
[2] Bartos, Matthew D., A. Mullapudi, and S. C. Troutman. "rrcf: Implementation of the Robust Random Cut Forest Algorithm for Anomaly Detection on Streams." Journal of Open Source Software 4, no. 35 (2019): 1336.
Version History
Introduced in R2023b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)