predict
Syntax
Description
predictedY = predict(Mdl,X)X using the trained quantile linear regression model
Mdl.
predictedY = predict(Mdl,X,Name=Value)
[
additionally returns a vector predictedY,crossingIndicator] = predict(___)crossingIndicator whose entries indicate
whether predictions for the specified quantiles cross each other.
Examples
Fit a quantile linear regression model using the 0.25, 0.50, and 0.75 quantiles.
Load the carbig data set, which contains measurements of cars made in the 1970s and early 1980s. Create a matrix X containing the predictor variables Acceleration, Displacement, Horsepower, and Weight. Store the response variable MPG in the variable Y.
load carbig
X = [Acceleration,Displacement,Horsepower,Weight];
Y = MPG;Delete rows of X and Y where either array has missing values.
R = rmmissing([X Y]); X = R(:,1:end-1); Y = R(:,end);
Partition the data into training data (XTrain and YTrain) and test data (XTest and YTest). Reserve approximately 20% of the observations for testing, and use the rest of the observations for training.
rng(0,"twister") % For reproducibility of the partition c = cvpartition(length(Y),"Holdout",0.20); trainingIdx = training(c); XTrain = X(trainingIdx,:); YTrain = Y(trainingIdx); testIdx = test(c); XTest = X(testIdx,:); YTest = Y(testIdx);
Train a quantile linear regression model. Specify to use the 0.25, 0.50, and 0.75 quantiles (that is, the lower quartile, median, and upper quartile). To improve the model fit, change the beta tolerance to 1e-6 instead of the default value 1e-4. Use a ridge (L2) regularization term of 1. Adjusting the regularization term can help prevent quantile crossing.
Mdl = fitrqlinear(XTrain,YTrain,Quantiles=[0.25,0.50,0.75], ...
BetaTolerance=1e-6,Lambda=1)Mdl =
RegressionQuantileLinear
ResponseName: 'Y'
CategoricalPredictors: []
ResponseTransform: 'none'
Beta: [4×3 double]
Bias: [17.0004 23.0029 29.5243]
Quantiles: [0.2500 0.5000 0.7500]
Properties, Methods
Mdl is a RegressionQuantileLinear model object. You can use dot notation to access the properties of Mdl. For example, Mdl.Beta and Mdl.Bias contain the linear coefficient estimates and estimated bias terms, respectively. Each column of Mdl.Beta corresponds to one quantile, as does each element of Mdl.Bias.
In this example, you can use the linear coefficient estimates and estimated bias terms directly to predict the test set responses for each of the three quantiles in Mdl.Quantiles. In general, you can use the predict object function to make quantile predictions.
predictedY = XTest*Mdl.Beta + Mdl.Bias
predictedY = 78×3
12.3963 16.2569 19.5263
5.8328 10.1568 12.6058
17.1726 20.6398 24.9748
23.3790 28.1122 31.3617
17.0036 22.5314 23.0539
16.6120 17.0713 20.1062
10.9274 12.3302 13.2707
14.9130 14.6659 12.7100
16.3103 17.7497 20.8477
19.6229 25.7109 30.5389
19.5583 24.6621 30.4345
12.9525 14.4508 16.0004
14.8525 16.1338 16.4112
24.1648 31.1758 33.9310
15.1039 17.8497 19.2013
⋮
isequal(predictedY,predict(Mdl,XTest))
ans = logical
1
Each column of predictedY corresponds to a separate quantile (0.25, 0.5, or 0.75).
Visualize the predictions of the quantile linear regression model. First, create a grid of predictor values.
minX = floor(min(X))
minX = 1×4
8 68 46 1613
maxX = ceil(max(X))
maxX = 1×4
25 455 230 5140
gridX = zeros(100,size(X,2)); for p = 1:size(X,2) gridp = linspace(minX(p),maxX(p))'; gridX(:,p) = gridp; end
Next, use the trained model Mdl to predict the response values for the grid of predictor values.
gridY = predict(Mdl,gridX)
gridY = 100×3
20.8073 25.4104 29.1436
20.6991 25.2907 29.0251
20.5909 25.1711 28.9066
20.4828 25.0514 28.7881
20.3746 24.9318 28.6696
20.2664 24.8121 28.5512
20.1583 24.6924 28.4327
20.0501 24.5728 28.3142
19.9419 24.4531 28.1957
19.8337 24.3335 28.0772
19.7256 24.2138 27.9587
19.6174 24.0941 27.8402
19.5092 23.9745 27.7217
19.4011 23.8548 27.6032
19.2929 23.7351 27.4848
⋮
For each observation in gridX, the predict object function returns predictions for the quantiles in Mdl.Quantiles.
View the gridY predictions for the second predictor (Displacement). Compare the quantile predictions to the true test data values.
predictorIdx = 2; plot(XTest(:,predictorIdx),YTest,".") hold on plot(gridX(:,predictorIdx),gridY(:,1)) plot(gridX(:,predictorIdx),gridY(:,2)) plot(gridX(:,predictorIdx),gridY(:,3)) hold off xlabel("Predictor (Displacement)") ylabel("Response (MPG)") legend(["True values","0.25 predicted values", ... "0.50 predicted values","0.75 predicted values"]) title("Test Data")
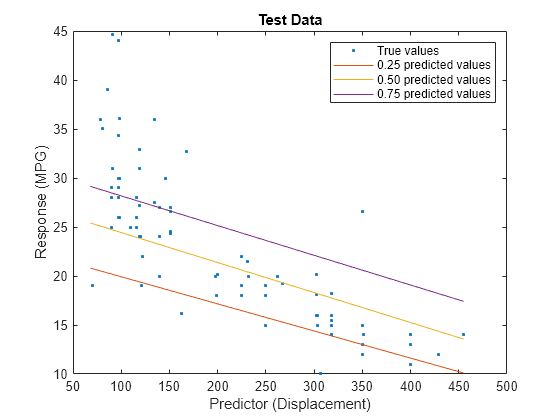
The red line shows the predictions for the 0.25 quantile, the yellow line shows the predictions for the 0.50 quantile, and the purple line shows the predictions for the 0.75 quantile. The blue points indicate the true test data values.
Notice that the quantile prediction lines do not cross each other.
When training a quantile linear regression model, you can use a ridge (L2) regularization term to prevent quantile crossing.
Load the carbig data set, which contains measurements of cars made in the 1970s and early 1980s. Create a table containing the predictor variables Acceleration, Cylinders, Displacement, and so on, as well as the response variable MPG.
load carbig cars = table(Acceleration,Cylinders,Displacement, ... Horsepower,Model_Year,Weight,MPG);
Remove rows of cars where the table has missing values.
cars = rmmissing(cars);
Partition the data into training and test sets using cvpartition. Use approximately 80% of the observations as training data, and 20% of the observations as test data.
rng(0,"twister") % For reproducibility of the data partition c = cvpartition(height(cars),"Holdout",0.20); trainingIdx = training(c); carsTrain = cars(trainingIdx,:); testIdx = test(c); carsTest = cars(testIdx,:);
Train a quantile linear regression model. Use the 0.25, 0.50, and 0.75 quantiles (that is, the lower quartile, median, and upper quartile). To improve the model fit, change the beta tolerance to 1e-6 instead of the default value 1e-4.
Mdl = fitrqlinear(carsTrain,"MPG",Quantiles=[0.25 0.5 0.75], ... BetaTolerance=1e-6);
Mdl is a RegressionQuantileLinear model object.
Determine if the test data predictions for the quantiles in Mdl.Quantiles cross each other by using the predict object function of Mdl. The crossingIndicator output argument contains a value of 1 (true) for any observation with quantile predictions that cross.
[~,crossingIndicator] = predict(Mdl,carsTest); sum(crossingIndicator)
ans = 2
In this example, two of the observations in carsTest have quantile predictions that cross each other.
To prevent quantile crossing, specify the Lambda name-value argument in the call to fitrqlinear. Use a 0.1 ridge (L2) penalty term.
newMdl = fitrqlinear(carsTrain,"MPG",Quantiles=[0.25 0.5 0.75], ... BetaTolerance=1e-6,Lambda=0.1); [predictedY,newCrossingIndicator] = predict(newMdl,carsTest); sum(newCrossingIndicator)
ans = 0
With regularization, the predictions for the test data set do not cross for any observations.
Visualize the predictions returned by newMdl by using a scatter plot with a reference line. Plot the predicted values along the vertical axis and the true response values along the horizontal axis. Points on the reference line indicate correct predictions.
plot(carsTest.MPG,predictedY(:,1),".") hold on plot(carsTest.MPG,predictedY(:,2),".") plot(carsTest.MPG,predictedY(:,3),".") plot(carsTest.MPG,carsTest.MPG) hold off xlabel("True MPG") ylabel("Predicted MPG") legend(["0.25 quantile values","0.50 quantile values", ... "0.75 quantile values","Reference line"], ... Location="southeast") title("Test Data")
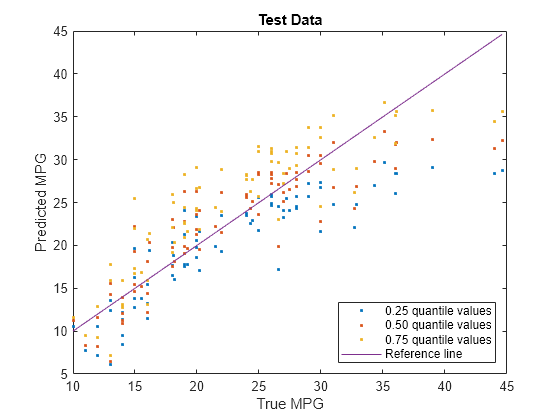
Blue points correspond to the 0.25 quantile, red points correspond to the 0.50 quantile, and yellow points correspond to the 0.75 quantile.
For a more in-depth example, see Regularize Quantile Regression Model to Prevent Quantile Crossing.
Input Arguments
Trained quantile linear regression model, specified as a RegressionQuantileLinear or CompactRegressionQuantileLinear model object.
Predictor data, specified as a numeric matrix or a table. Unless you specify the
ObservationsIn name-value argument, each row of
X corresponds to one observation, and each column corresponds to
one variable.
For a numeric matrix:
The variables in
Xmust have the same order as the predictor variables that trainedMdl.If you train
Mdlusing a table (for example,Tbl) andTblcontains only numeric predictor variables, thenXcan be a numeric matrix. To treat numeric predictors inTblas categorical during training, identify categorical predictors by using theCategoricalPredictorsname-value argument offitrqlinear. IfTblcontains heterogeneous predictor variables (for example, numeric and categorical data types) andXis a numeric matrix, thenpredictissues an error.
For a table:
predictdoes not support multicolumn variables or cell arrays other than cell arrays of character vectors.If you train
Mdlusing a table (for example,Tbl), then all predictor variables inXmust have the same variable names and data types as the variables that trainedMdl(stored inMdl.PredictorNames). However, the column order ofXdoes not need to correspond to the column order ofTbl. Also,TblandXcan contain additional variables (response variable, observation weights, and so on), butpredictignores them.If you train
Mdlusing a numeric matrix, then the predictor names inMdl.PredictorNamesmust be the same as the corresponding predictor variable names inX. To specify predictor names during training, use thePredictorNamesname-value argument offitrqlinear. All predictor variables inXmust be numeric vectors.Xcan contain additional variables (response variable, observation weights, and so on), butpredictignores them.
If you set Standardize to true in
fitrqlinear when training Mdl, then the
software standardizes the numeric columns of the predictor data using the corresponding
means (Mdl.Mu) and standard deviations
(Mdl.Sigma).
Note
If you orient your predictor matrix so that observations correspond to columns and
specify ObservationsIn="columns", then you might experience a
significant reduction in computation time. You cannot specify
ObservationsIn="columns" for predictor data in a table.
Data Types: single | double | table
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: predict(Mdl,X,ObservationsIn="columns") specifies that
columns in the predictor data correspond to observations.
Quantiles for which to compute predictions, specified as a vector of values in
Mdl.Quantiles. The predict function returns
predictions for each quantile.
Example: Quantiles=[0.4 0.6]
Data Types: single | double | char | string
Predictor data observation dimension, specified as "rows" or
"columns".
Note
If you orient your predictor matrix so that observations correspond to columns
and specify ObservationsIn="columns", then you might experience a
significant reduction in computation time. You cannot specify
ObservationsIn="columns" for predictor data in a table.
Example: ObservationsIn="columns"
Data Types: char | string
Since R2025a
Predicted response value to use for observations with missing predictor values,
specified as "quantile", a numeric scalar, or a numeric
vector.
| Value | Description |
|---|---|
"quantile" | predict uses the specified quantile of the
observed response values in the training data as the predicted response value
for observations with missing predictor values. |
| Numeric scalar or vector |
|
Example: PredictionForMissingValue=NaN
Data Types: single | double | char | string
Output Arguments
Quantile crossing indicator, returned as a logical vector. Each entry corresponds to
an observation in X. A value of 1
(true) indicates that the corresponding observation has predictions
that cross. That is, two quantiles q1 and q2 exist
in Quantiles such that q1 < q2
and predictedYq1
>
predictedYq2.
Version History
Introduced in R2024bYou can predict responses for a compact quantile regression model (CompactRegressionQuantileLinear).
You can also specify the prediction values for observations with missing predictor
values by using the PredictionForMissingValues name-value argument. In previous releases,
observations with missing predictor values have NaN predictions. That is,
the behavior is equivalent to PredictionForMissingValue=NaN.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)