Train Decision Trees Using Classification Learner App
This example shows how to create and compare various classification trees using Classification Learner, and export trained models to the workspace to make predictions for new data.
You can train classification trees to predict responses to data. To predict a response, follow the decisions in the tree from the root (beginning) node down to a leaf node. The leaf node contains the response.
Statistics and Machine Learning Toolbox™ trees are binary. Each step in a prediction involves checking the value of one predictor (variable). For example, here is a simple classification tree:

This tree predicts classifications based on two predictors, x1 and
x2. To predict, start at the top node. At each decision, check
the values of the predictors to decide which branch to follow. When the branches reach a
leaf node, the data is classified either as type 0 or
1.
This example uses Fisher's 1936 iris data. The iris data contains measurements of flowers: the petal length, petal width, sepal length, and sepal width for specimens from three species. Train a classifier to predict the species based on the predictor measurements.
In MATLAB®, load the
fisheririsdata set and create a table of measurement predictors (or features) using variables from the data set to use for a classification.fishertable = readtable("fisheriris.csv");On the Apps tab, in the Machine Learning and Deep Learning group, click Classification Learner.
On the Learn tab, in the File section, click New Session > From Workspace.
In the New Session from Workspace dialog box, select the table
fishertablefrom the Data Set Variable list (if necessary).Observe that the app has selected response and predictor variables based on their data type. Petal and sepal length and width are predictors, and species is the response that you want to classify. For this example, do not change the selections.

To accept the default validation scheme and continue, click Start Session. The default validation option is cross-validation, to protect against overfitting.
Classification Learner creates a scatter plot of the data.

Use the scatter plot to investigate which variables are useful for predicting the response. To visualize the distribution of species and measurements, select different variables in the X and Y lists under Predictors to the right of the plot. Observe which variables separate the species colors most clearly.
Observe that the
setosaspecies (blue points) is easy to separate from the other two species with all four predictors. Theversicolorandvirginicaspecies are much closer together in all predictor measurements, and overlap especially when you plot sepal length and width.setosais easier to predict than the other two species.Train fine, medium, and coarse trees simultaneously. The Models pane already contains a fine tree model. Add medium and coarse tree models to the list of draft models. On the Learn tab, in the Models section, click the arrow to open the gallery. In the Decision Trees group, click Medium Tree. The app creates a draft medium tree in the Models pane. Reopen the model gallery and click Coarse Tree in the Decision Trees group. The app creates a draft coarse tree in the Models pane.
In the Train section, click Train All and select Train All. The app trains the three tree models.
Note
If you have Parallel Computing Toolbox™, then the Use Parallel button is selected by default. After you click Train All and select Train All or Train Selected, the app opens a parallel pool of workers. During this time, you cannot interact with the software. After the pool opens, you can continue to interact with the app while models train in parallel.
If you do not have Parallel Computing Toolbox, then the Use Background Training check box in the Train All menu is selected by default. After you select an option to train models, the app opens a background pool. After the pool opens, you can continue to interact with the app while models train in the background.
Note
Validation introduces some randomness into the results. Your model validation results can vary from the results shown in this example.
In the Models pane, each model has a validation accuracy score that indicates the percentage of correctly predicted responses. The app highlights the highest Accuracy (Validation) value (or values) by outlining it in a box.
Click a model to view the results, which are displayed in the Summary tab. To open this tab, right-click the model and select Summary.
For each model, examine the scatter plot. On the Learn tab, in the Plots and Results section, click the arrow to open the gallery, and then click Scatter in the Validation Results group. An X indicates misclassified points.
For all three models, the blue points (
setosaspecies) are all correctly classified, but some of the other two species are misclassified. Under Plot, switch between the Data and Model Predictions options. Observe the color of the incorrect (X) points. Alternatively, while plotting model predictions, to view only the incorrect points, clear the Correct check box.To try to improve the models, include different features during model training. See if you can improve the model by removing features with low predictive power.
On the Learn tab, in the Options section, click Feature Selection.
In the Default Feature Selection tab, you can select different feature ranking algorithms to determine the most important features. After you select a feature ranking algorithm, the app displays a plot of the sorted feature importance scores, where larger scores (including
Infs) indicate greater feature importance. The table shows the ranked features and their scores.In this example, the Chi2, ReliefF, ANOVA, and Kruskal Wallis feature ranking algorithms all identify the petal measurements as the most important features. Under Feature Ranking Algorithm, click Chi2.
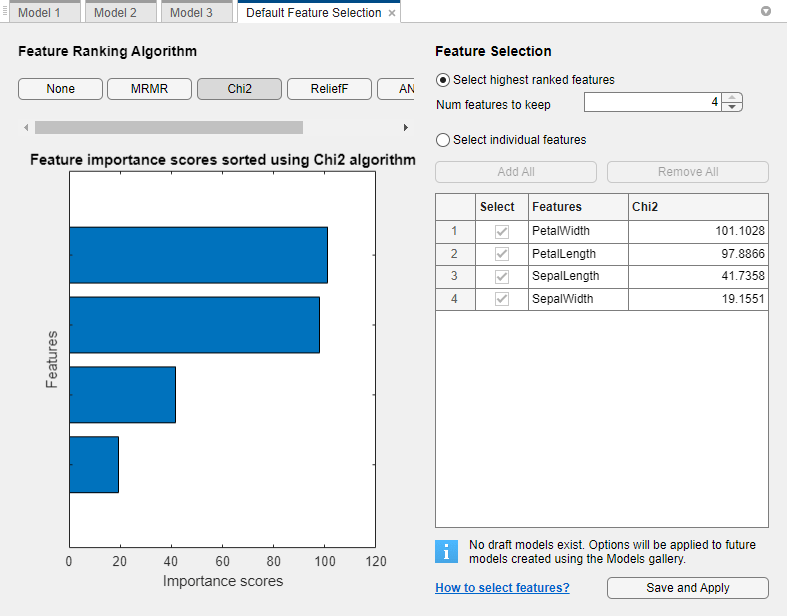
Under Feature Selection, use the default option of selecting the highest ranked features to avoid bias in the validation metrics. Specify to keep 2 of the 4 features for model training. Click Save and Apply. The app applies the feature selection changes to new models created using the Models gallery.
Train new tree models using the reduced set of features. On the Learn tab, in the Models section, click the arrow to open the gallery. In the Decision Trees group, click All Trees. In the Train section, click Train All and select Train All or Train Selected.
The models trained using only two measurements perform comparably to the models containing all predictors. The models predict no better using all the measurements compared to only the two measurements. If data collection is expensive or difficult, you might prefer a model that performs satisfactorily without some predictors.
Note the last model in the Models pane, a Coarse Tree model trained using only 2 of 4 predictors. The app displays how many predictors are excluded. To check which predictors are included, click the model in the Models pane, and note the check boxes in the expanded Feature Selection section of the model Summary tab.
Note
If you use a cross-validation scheme and choose to perform feature selection using the Select highest ranked features option, then for each training fold, the app performs feature selection before training a model. Different folds can select different predictors as the highest ranked features. The table on the Default Feature Selection tab shows the list of predictors used by the full model, trained on the training and validation data.
Train new tree models using another subset of measurements. On the Learn tab, in the Options section, click Feature Selection. In the Default Feature Selection tab, click MRMR under Feature Ranking Algorithm. Under Feature Selection, specify to keep 3 of the 4 features for model training. Click Save and Apply.
On the Learn tab, in the Models section, click the arrow to open the gallery. In the Decision Trees group, click All Trees. In the Train section, click Train All and select Train All or Train Selected.
The models trained using only 3 of 4 predictors do not perform as well as the other trained models.
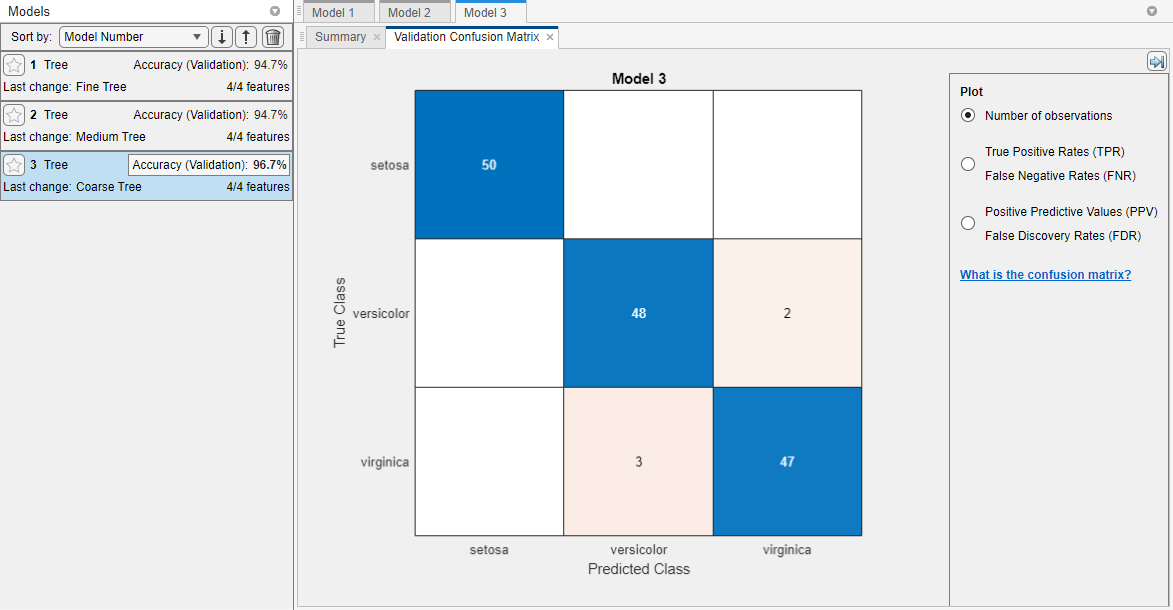
Choose a best model among those of similar accuracy by examining the performance in each class. For example, select the coarse tree that includes 2 of 4 predictors. Inspect the accuracy of the predictions in each class. On the Learn tab, in the Plots and Results section, click the arrow to open the gallery, and then click Confusion Matrix (Validation) in the Validation Results group. Use this plot to understand how the currently selected classifier performed in each class. View the matrix of true class and predicted class results.
Look for areas where the classifier performed poorly by examining cells off the diagonal that display high numbers and are red. In these red cells, the true class and the predicted class do not match. The data points are misclassified.

In this figure, examine the third cell in the middle row. In this cell, true class is
versicolor, but the model misclassified the points asvirginica. For this model, the cell shows 2 misclassified (your results can vary). To view percentages instead of numbers of observations, select the True Positive Rates option under Plot controls.You can use this information to help you choose the best model for your goal. If false positives in this class are very important to your classification problem, then choose the best model at predicting this class. If false positives in this class are not very important, and models with fewer predictors do better in other classes, then choose a model to tradeoff some overall accuracy to exclude some predictors and make future data collection easier.
Compare the confusion matrix for each model in the Models pane. Check the Feature Selection section of the model Summary tab to see which predictors are included in each model.
In this example, the coarse tree that includes 2 of 4 predictors performs as well as the coarse tree with all predictors. That is, both models provide the same validation accuracy and have the same confusion matrix.
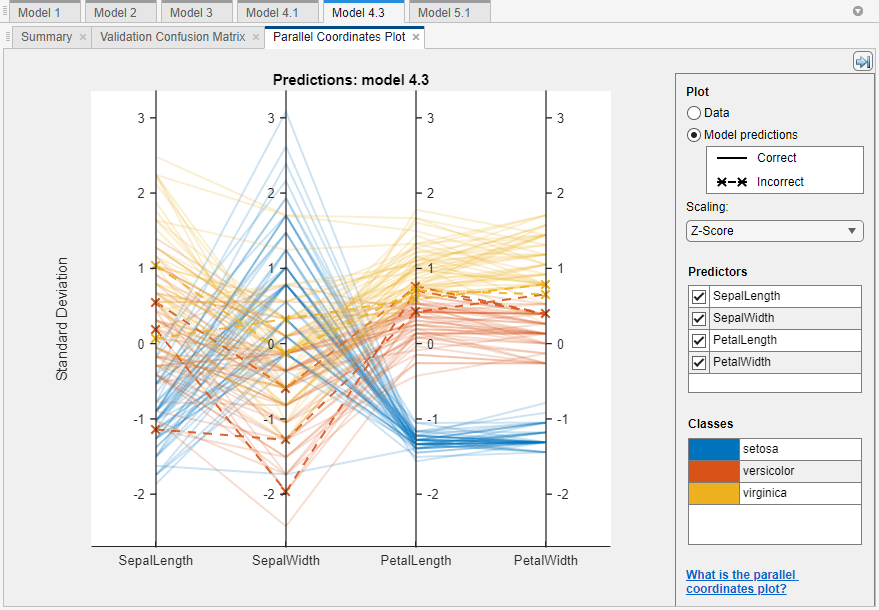
To further investigate features to include or exclude, use the parallel coordinates plot. On the Learn tab, in the Plots and Results section, click the arrow to open the gallery, and then click Parallel Coordinates in the Validation Results group. You can see that petal length and petal width are the features that separate the classes best.
To learn about model hyperparameter settings, choose a model in the Models pane and expand the Model Hyperparameters section in the model Summary tab. Compare the coarse and medium tree models, and note the differences in the model hyperparameters. In particular, the Maximum number of splits setting is 4 for coarse trees and 20 for medium trees. This setting controls the tree depth.
To try to improve the coarse tree model further, change the Maximum number of splits setting. First, click the model in the Models pane. Right-click the model and select Duplicate. In the Summary tab, change the Maximum number of splits value. Then, in the Train section of the Learn tab, click Train All and select Train Selected.
Click on the best trained model in the Models pane.
You can export a full or compact version of the trained model to the workspace. On the Learn tab, in the Export section, click Export Model and select Export Model to Workspace. To exclude the training data and export a compact model, clear the check box in the Export Classification Model dialog box. You can still use the compact model for making predictions on new data. In the dialog box, click OK to accept the default variable name. Look in the command window to see information about the results.
To visualize your decision tree model, enter:
view(trainedModel.ClassificationTree,"Mode","graph")
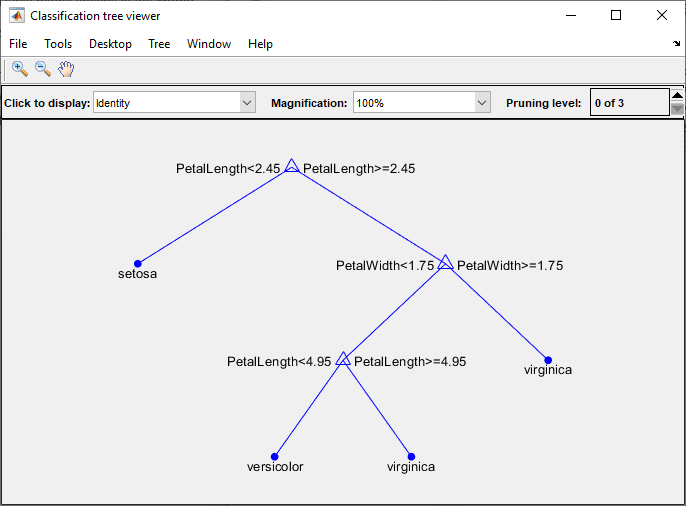
You can use the exported classifier to make predictions on new data. For example, to make predictions for the
fishertabledata in your workspace, enter:The output[yfit,scores] = trainedModel.predictFcn(fishertable)
yfitcontains a class prediction for each data point. The outputscorescontains the class scores returned by the trained model.scoresis an n-by-k array, where n is the number of data points and k is the number of classes in the trained model.If you want to automate training the same classifier with new data, or learn how to programmatically train classifiers, you can generate code from the app. To generate code for the best trained model, on the Learn tab, in the Export section, click Generate Function.
The app generates code from your model and displays the file in the MATLAB Editor. To learn more, see Generate MATLAB Code to Train Model with New Data.
Use the same workflow to evaluate and compare the other classifier types you can train in Classification Learner.
To try all the nonoptimizable classifier model presets available for your data set:
On the Learn tab, in the Models section, click the arrow to open the gallery of classification models.
In the Get Started group, click All.
In the Train section, click Train All and select Train All.
To learn about other classifier types, see Train Classification Models in Classification Learner App.
See Also
Topics
- Train Classification Models in Classification Learner App
- Select Data for Classification or Open Saved App Session
- Choose Classifier Options
- Feature Selection and Feature Transformation Using Classification Learner App
- Visualize and Assess Classifier Performance in Classification Learner
- Export Classification Model to Predict New Data