稀疏矩阵的图形表示
此示例说明 NASA 翼型的有限元网格,包括两个尾翼。有关翼型发展历史的详细信息,请访问 NACA Airfoils (nasa.gov)。
数据存储在文件 airfoil.mat 中。数据由 4253 对 (x,y) 网格点坐标组成。此外,还包含一个由 12,289 对索引 (i,j) 组成的数组,这些索引指定网格点之间的连接。
将数据文件加载到工作区。
load airfoil查看有限元网格
首先,将 x 和 y 分别以 为比例缩小,使其处于范围 内。然后,从 (i,j) 连接点构造稀疏邻接矩阵,并将其设为正定矩阵。最后,使用 (x,y) 作为顶点(网格点)的坐标绘制邻接矩阵。
% Scaling x and y x = pow2(x,-32); y = pow2(y,-32); % Forming the sparse adjacency matrix and making it positive definite n = max(max(i),max(j)); A = sparse(i,j,-1,n,n); A = A + A'; d = abs(sum(A)) + 1; A = A + diag(sparse(d)); % Plotting the finite element mesh gplot(A,[x y])
ans = 86493×1
0
0
NaN
0.0002
0
NaN
0
0.0002
NaN
0.0002
0.0002
NaN
0.0054
0.0002
NaN
⋮
title('Airfoil Cross-Section')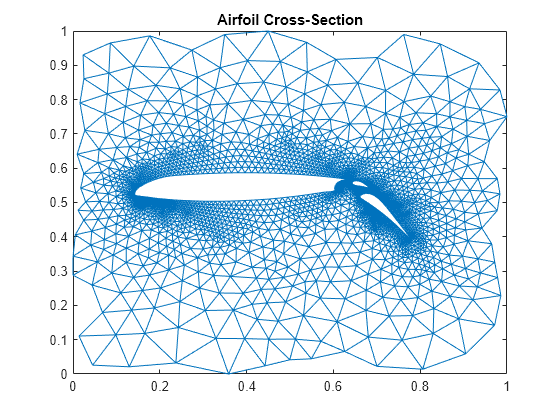
可视化稀疏模式
您可以使用 spy 可视化矩阵中的非零元素,该函数尤其适用于查看稀疏矩阵中的稀疏模式。spy(A) 可以绘制矩阵 A 的稀疏模式。
spy(A)
title('Airfoil Adjacency Matrix')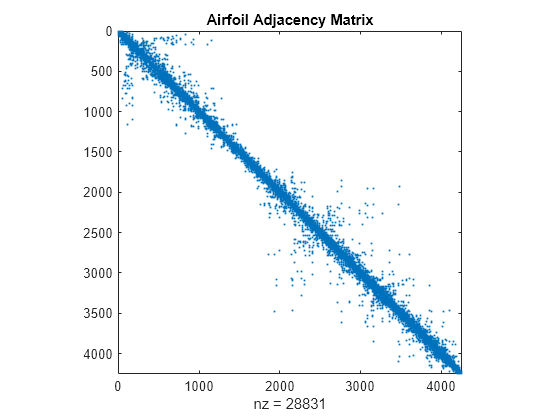
对称重新排序 - 反向卡西尔-麦基
symrcm 使用反向卡西尔-麦基方法对邻接矩阵重新排序。r = symrcm(A) 返回置换向量 r,因此,相对于 A,A(r,r) 往往具有更接近对角线的对角线元素。对于来自“瘦长”问题的矩阵的 LU 或乔列斯基分解来说,这是一种很好的预排序方法。此方法对于对称和非对称矩阵都适用。
r = symrcm(A);
spy(A(r,r))
title('Reverse Cuthill-McKee')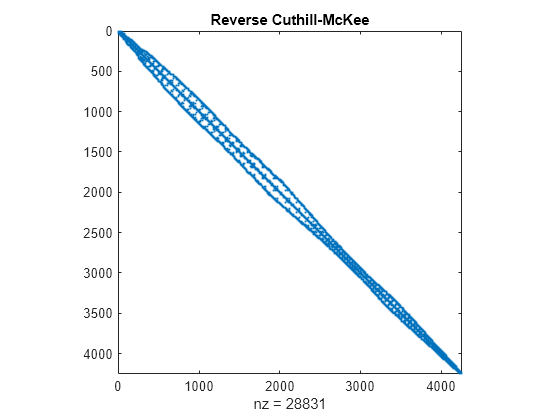
对称重新排序 - 列置换
使用 j = COLPERM(A) 可返回一个置换向量,该向量以非零计数的非降序对稀疏矩阵 A 的各列进行重新排序。此方法有时很有用,可作为对 LU 分解的预排序方法,如 lu(A(:,j))。
j = colperm(A);
spy(A(j,j))
title('Column Count Reordering')
对称重新排序 - 对称近似最小度
symamd 可以进行对称近似最小度置换。对于对称正定矩阵 A,命令 p = symamd(S) 返回置换向量 p,因此 S(p,p) 倾向于比 S 具有更稀疏的乔列斯基因子。有时 symamd 也适用于对称的非正定矩阵。
m = symamd(A);
spy(A(m,m))
title('Approximate Minimum Degree')