framelbl
Syntax
Description
Examples
Partition a categorical label sequence into frames of length 4.
catLbl = ["healthy" "healthy" "sick" ... "sick" "healthy" "healthy" ... "healthy" "sick" "sick" "sick"]; framedLabels = framelbl(catLbl,4)
framedLabels = 4×2 string
"healthy" "healthy"
"healthy" "healthy"
"sick" "healthy"
"sick" "sick"
Partition a single label defined as a categorical input into frames using name-value arguments.
Define a categorical scalar Y.
lblName = categorical("Y")lblName = categorical
Y
Note that this label definition has one element, and its datatype is categorical.
To frame a scalar label, either:
Set the input label mode to
"attribute"and specify a signal length.Repeat the scalar label. In this case, the input label mode must be
"mask"(default value) and you must not specify a signal length.
Partition a categorical scalar into four-sample frames. Set the input label mode to "attribute" with a signal length of 15 samples. Use an underlap length of 2 samples, an initial index of 3. Specify the incomplete frame rule to "padwithmissing".
fl = 4; sl = 15; ul = 2; ii0 = 3; flblAttr = framelbl(lblName,fl, ... InputLabelMode="attribute",SignalLength=sl,... UnderlapLength=ul,InitialIndex=ii0, ... IncompleteFrameRule="padwithmissing")
flblAttr = 4×3 categorical
Y Y Y
Y Y <undefined>
Y Y <undefined>
Y Y <undefined>
The output frame label sequence follows the framing specifications in this order:
The
framelblfunction reads the categorical scalar inputlblName, with the input label mode"attribute", and repeatsY15 times.The framing starts with the third sample, following the value of
InitialIndexspecified in the function call.Then, the first frames contains the samples 3, 4, 5, and 6. The framing operation skips the samples 7 and 8, as per the
UnderlapLengthspecification.The second frame includes samples 9 through 12. The
framelblfunction skips samples 13 and 14.The third frame includes the sample 15 and gets completed with missing-name values. Since
lblNameis categorical, then the frame completes with<undefined>values.
Define a label sequence by repeating a categorical scalar up to sl samples. Partition a categorical scalar into frames. Specify the frame length, underlap length, initial index, and incomplete frame rule. Set the input label mode to "mask".
lblSeq = repmat(lblName,sl,1); flblMask = framelbl(lblSeq,fl, ... InputLabelMode="mask", ... UnderlapLength=ul,InitialIndex=ii0, ... IncompleteFrameRule="padwithmissing")
flblMask = 4×3 categorical
Y Y Y
Y Y <undefined>
Y Y <undefined>
Y Y <undefined>
Framing a categorical scalar declared as "attribute" with a signal length yields the same results as framing a categorical array that explicitly lists the category signal.
Define a categorical sequence, two categories, and a category priority list. Partition a categorical sequence.
Consider a categorical sequence defined by the 18-by-2 logical matrix logMat. The first and second columns correspond to categories named "A" and "B", respectively.
logMat = logical([ ... 0 0 1 1 1 0 0 1 1 0 0 0 1 1 1 1 0 1; ... 1 1 0 0 0 1 1 1 1 0 1 1 0 1 0 1 1 0]'); catList = ["A" "B"];
In each row, the associated category is "A" if 1 is listed in the first column, or "B" if 1 is listed in the second column. If none of the columns lists 1, the corresponding category is <undefined>. For multiple instances of 1 listed, the priority list of categories defines the category to associate with each row.
Category "B" has higher priority than "A". Define a category priority list.
catPriority = ["B" "A"];
Partition the categorical sequence defined by the logical matrix into frames of length 7 with 3 samples of overlap between adjoining frames. Use the categories "A" and "B".
[framedLabels,finalCond] = framelbl(logMat,7, ... OverlapLength=3,Categories=catList, ... PriorityList=catPriority)
framedLabels = 7×3 categorical
B A B
B B <undefined>
A B B
A B B
A B A
B <undefined> B
B B A
finalCond = 6×2 logical array
1 0
0 1
1 0
0 1
0 1
1 0
Define an ROI table. Partition label sequence into frames. Consolidate label frames.
Specify five ranges between 5 and 20, and define the categories "A", "B", "C", "D", and "E". Let the variables minimums and maximums be the minimum and maximum values that correspond to each range associated with the categories. The value 12 associates with the categories "C" and "D", in which case framelbl enables labeling based on a priority order.
minimums = [5 10 12 15 18]'; maximums = [9 12 14 17 20]'; categories = ["E" "D" "C" "B" "A"]';
Define an ROI table with the numeric ranges and category labels. Specify the variable names as "Range" and "Category", respectively.
ROItable = table([minimums maximums],categories,... VariableNames=["Range" "Category"])
ROItable=5×2 table
Range Category
________ ________
5 9 "E"
10 12 "D"
12 14 "C"
15 17 "B"
18 20 "A"
Partition an ROI table into frames of length 6 samples.
fl = 6; [flbl,fnlcond,fnlidx] = framelbl(ROItable,fl)
flbl = 6×3 string
<missing> "E" "C"
<missing> "E" "C"
<missing> "E" "B"
<missing> "D" "B"
"E" "D" "B"
"E" "C" "A"
fnlcond=1×2 table
Range Category
________ ________
19 20 "A"
fnlidx = 1
The outputs from the framing operation show the following:
Since the ROI table limits range from 5 to 20, thus not having values from 1 to 4, the framing operation completes the first four labels of
flblwith<missing>values.The framing operation assigns
"C"to the last sample of the second frame because it has a higher priority over"D", from the default lexicographical priority order.
Partition an ROI table into frames. Consolidate the framed label sequence with the most frequent label in each frame.
flblGroup = framelbl(ROItable,fl,ConsolidationMethod="mode")flblGroup = 1×3 string
"E" "E" "B"
The most frequent grade label in each frame of flbl are "E", "E" and "B", which yield the same result as flblGroup.
Load a signal and its associated label sequence data. Create datastores and frame signal and labels. Combine, read, and show the framed signal and labels.
Load the file framedata, which contains a signal vector and its associated label sequence. The vector is a random audio-like signal with a length L of 100,000 samples at a sampling rate of 16 kHz. The label sequence is a categorical vector where "S" corresponds to speech samples and "N" corresponds to no speech.
load framedataCreate the datastores sigAds and lblAds from the arrays sig and lbl. Read L samples of data along the second dimension in each iteration.
sigAds = arrayDatastore(sig,IterationDimension=2,ReadSize=L); lblAds = arrayDatastore(lbl,IterationDimension=2,ReadSize=L);
Partition both signal and label sequence into frames of length 1024 with 512 samples of overlap between adjoining frames. Set the ConsolidationMethod property to "mode" for the label framing operation to assign the most frequent category in each framed label. Use the transform function to transform the datastores.
transformedSigDS = transform(sigAds,... @(x){framesig(x{:},1024,OverlapLength=512)}); transformedLblDS = transform(lblAds,... @(x){framelbl(x{:},1024,OverlapLength=512, ... ConsolidationMethod="mode")});
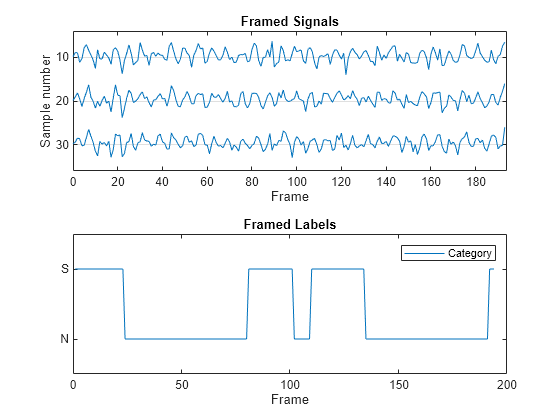
Combine the transformed datastores coming from the signal and label framing operations. Read and plot the resulting datastore frames, showing the samples 10, 20, and 30 of the framed signal data, and the labeled category in each frame.
combinedDS = combine(transformedSigDS,transformedLblDS); framedSignalAndLabel = read(combinedDS); tiledlayout flow nexttile strips(framedSignalAndLabel{1}(10:10:30,:)') title("Framed Signals") xlabel("Frame") ylabel("Sample number") yticklabels([30 20 10]) nexttile plot(framedSignalAndLabel{2}) title("Framed Labels") xlabel("Frame") legend("Category")
Label a speech signal. Extract time-domain features by frames. Partition a numeric label sequence into frames and consolidate values.
Load a data file containing a speech vector signal sampled at a frequency rate fs of 16 kHz. Define a labeling vector labels as a numeric vector describing the ratio between the signal amplitudes over the mean amplitude, rounding to the nearest integer.
[signal,fs] = audioread("MaleVolumeUp-16-mono-6secs.ogg");
amp = abs(signal);
labels = int8(amp/mean(amp));Set a frame length to 4096 samples. Extract time-domain features to a signal vector.
fl = 4096;
sFE = signalTimeFeatureExtractor(FrameSize=fl,SampleRate=fs, ...
ShapeFactor=true,RMS=true,CrestFactor=true);
fFeatures = extract(sFE,signal);Partition a numeric vector of labels into frames. Consolidate the label in each frame as the mean of the label values.
fLabels = framelbl(labels,fl,ConsolidationMethod="mean");Plot the signal and the numeric labels. Plot the consolidated frame labels and the normalized time-domain features in each frame.
tiledlayout vertical nexttile plot(signal) xline(fl:fl:length(signal),":") xlim([0 fl*ceil(length(signal)/fl)]) xlabel("Sample number") title("Signal") nexttile stairs(labels) xline(fl:fl:length(labels),":") xlim([0 fl*ceil(length(labels)/fl)]) xlabel("Sample number") title("Labels")
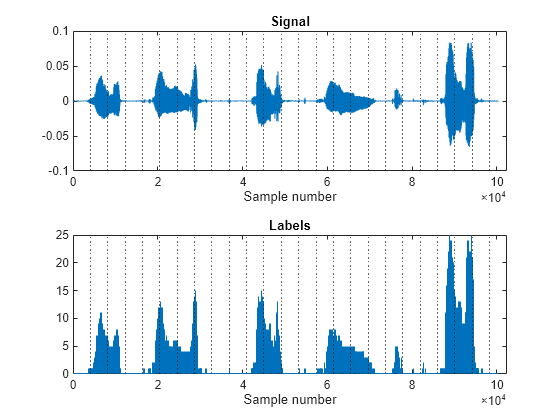
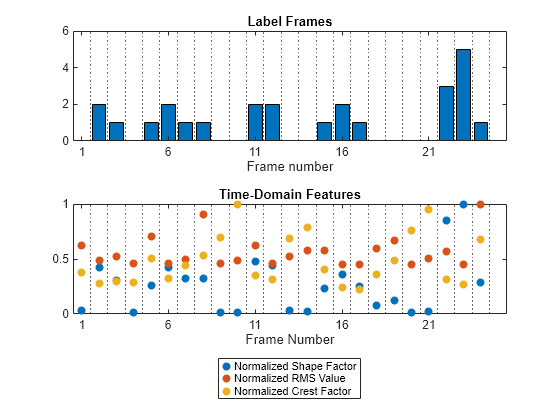
figure tiledlayout vertical nexttile b = bar((1:length(fLabels))-0.5,fLabels,FaceColor="flat"); xline(1:length(fLabels),":") xlim([0 ceil(length(labels)/fl)]) xticks((0:5:length(fLabels))+0.5) xticklabels((0:5:length(fLabels))+1) xlabel("Frame number") title("Label Frames") nexttile scatter(0.5:numel(fLabels)-0.5,fFeatures./max(fFeatures),"filled") xline(1:length(fLabels),":") xlabel("Frame Number") legend("Normalized "+["Shape Factor" "RMS Value" "Crest Factor"], ... Location="southoutside") title("Time-Domain Features") xlim([0 ceil(length(labels)/fl)]) xticks((0:5:length(fLabels))+0.5) xticklabels((0:5:length(fLabels))+1) box on
Input Arguments
Input signal, specified as a categorical vector, string array, numeric array, logical matrix, or table.
If
xis a string array, numeric array or logical matrix,framelblreturns anfl-by-NF matrixflbl, where NF is the number of frames.If
xis a logical matrix with P columns,framelblinterprets each column as a mask with true elements marking regions of interest (ROI) for each of P different categories,"1","2", ...,"P", and converts the logical matrix into a categorical label sequence. Each row of the matrix is an observation and each column of the matrix corresponds to a category. Theframelblfunction treats a logical row vector as a single observation and a logical column vector as a single category. If there are multiple categories in overlapping regions of interest, the function resolves the ambiguity by giving the highest priority to category"1"and the lowest priority to category"P". Finally,framelblbreaks the label sequence into equal-length frames.If
xis a table representing ROIs, this ROI table must contain two variables. The first variable is a two-column matrix such that each row defines the limits of a region of interest. The second variable contains the label values of the regions, specified as a categorical vector or a string array.framelblconverts the ROI table into a categorical or string sequence depending of the type specified for the label values. If there are overlapping regions of interest with different label values in the ROI table,framelblgives precedence to the label that appears first in lexicographic order. Finally,framelbldivides the label sequence into frames.
Example: ["current" "past" "past" "current"
"pass"].
Example: logical([0 0 1 0 1 0]').
Example: table([2 12; 7 16; 3 8],["U" "S" "S"]').
Note
If x is either an ROI table or a logical matrix, unlabeled
regions are labeled with missing values. For label values of categorical and string
types, missing values are <undefined> and
<missing>, respectively.
Data Types: categorical | string | table | double | single | logical | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Frame length, specified in samples as a positive integer scalar.
fl must be smaller than or equal to the length of input
x.
Example: fl = 5.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: framelbl("p",5,SignalLength=12,InputLabelMode="attribute",IncompleteFrameRule="padwithmissing")
partitions a 12-sample label sequence formed with the string "p" into
frames of length 5 using the "attribute" input label
mode and the "padwithmissing" incomplete frame rule as name-value pair
arguments.
Input label mode, specified as either "mask" or
"attribute".
"mask"—framelblassumes that each element in the label sequence corresponds to a signal sample."attribute"—framelblassumes that the inputxcorresponds to all samples in the data sequence. In this case, the input must be a categorical, string, or numeric scalar andframelblreturns anfl-by-NF matrixflblwith all entries equal tox, where NF is the number of frames. A logical1corresponds to a label category"1"and a logical0corresponds to an<undefined>label.
Data Types: char | string
Length of the signal associated to the input label sequence, specified as a
positive integer scalar. SignalLength must be greater than the
frame length fl.
If InputLabelMode is:
"attribute"— You must specifySignalLength."mask"—SignalLengthcan be specified only ifxis an ROI table. Then:If
SignalLengthfalls within the specified ROI limits, the function ignores the regions beyondSignalLength.If
SignalLengthexceeds the maximum ROI limit, the function padsxwith<missing>values.If
SignalLengthis not specified, the function infersSignalLengthfrom the input ROI table and sets it to the maximum index in the ROI limit matrix.
Sample rate of the signal associated to the input label sequence, specified as a
positive scalar. SampleRate can be specified only if
x is an ROI table.
If
SampleRateis specified,framelblassumes that the table contains region limits in seconds. Prior to performing framing operations,framelblconverts time values to sample indices, rounding to the nearest integer.If
SampleRateis not specified, all region limits must be specified as positive integer indices.
Data Types: single | double
List of category names, specified as a string array.
Categories can be specified only if x is a
logical matrix.
The
framelblfunction interprets the ith column ofxas a binary mask corresponding to the ith category name inCategories.If
xis a logical matrix andCategoriesis not specified,framelblsets categories to the string array["1" "2" ... "P"].For other non-numeric values of
x, the value ofCategoriesis directly inferred fromx.
Data Types: string
Frame labeling consolidation method, specified as either
"none", "mode", "max",
"median", "mean", or
"priority". ConsolidationMethod specifies
the method to consolidate all the label values in a frame into a single value to
assign as the label of the frame. In this case, the output label sequence
flbl becomes a vector of frame labels.
framelbl performs the label consolidation depending of the
specified value of ConsolidationMethod:
"none"—framelbldoes not consolidate labels."mode"—framelblassigns the most frequently occurring label value in the frame (the mode) as the frame label."max"—framelblassigns the maximum label value in the frame as the frame label."median"—framelblassigns the median of label values in the frame as the frame label."mean"—framelblaverages the label values in a frame and specifies the average as the frame label."priority"—framelbluses the priority list given inPriorityListto consolidate label values. If the first element of the priority list is not present, the function specifies as frame label the first value in the priority list of which there is at least one instance in the frame.
Note
You can set ConsolidationMethod to certain values based
on the input type of x:
categorical or string —
ConsolidationMethodcan be only"mode"or"priority". This rule applies if you explicitly definexas a categorical sequence or a string array, or implicitly, by an ROI table or a logical matrix. For ordinal categorical inputs,ConsolidationMethodcan also be set to"median"or"max".numeric —
ConsolidationMethodcan take any value except"priority".
Data Types: char | string
Category priority list, specified as a categorical array, a string array, or a
numeric array. PriorityList specifies a vector of unique
categorical or string elements, or index of unique labels, ordered by the priority
with which they should be treated when regions with different label values overlap or
when consolidating frames that contain different label values.
PriorityListmust contain or point to all unique label values.framelblprioritizes label values following the order of the elements inPriorityList. The first value inPriorityListhas the highest priority.You can set a value for
PriorityListonly whenxis an ROI table or a logical array, or when you setConsolidationMethodto"priority".If
xis an ordinal categorical vector andConsolidationMethodis"priority",PriorityListoverrides the mathematical ordering ofx.If you do not specify
PriorityList,framelblassigns a value by default. Ifxis of typelogical,PriorityListgets it value fromCategories. Otherwise,PriorityListdefaults to a lexicographically ordered list of label values fromx.
Data Types: categorical | string | double | single | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Overlap length, specified as a nonnegative integer scalar. OverlapLength
is the number of overlap samples between adjoining frames.
OverlapLength must be nonnegative, smaller than the frame
length, and applies only when UnderlapLength is not
specified.
Underlap length, specified as a nonnegative integer scalar.
UnderlapLength is the number of samples to be skipped after
each frame, thus reducing the frame rate. UnderlapLength must be
nonnegative and applies only when OverlapLength is not
specified.
Array of initial conditions, specified as a categorical vector, string array,
numeric array, logical matrix, or table. InitialCondition
specifies an initial condition that is stacked on top of the input label sequence
x.
InitialConditionmust be in such shape that it can be concatenated withx.InitialConditionmust have the same data type asx.If
InputLabelModeis set to"attribute",InitialConditionmust be empty.If
xis an ROI table, the initial condition must also be an ROI table whose ROI limits do not overlap with the ROI limits in the input ROI table.
Example: framelbl(["S" "S" "N" "N" "S"],3,InitialCondition=["IC"
"IC"]) partitions a 5-sample string vector into frames with a length of
three samples. The initial condition is also a string vector.
Example: framelbl([1 1 0 1 0 0 0],5,InitialCondition=[0 0 1])
partitions a 7-sample logical vector into frames with a length of five samples. The
initial condition is also a logical vector.
Data Types: categorical | string | table | double | single | logical | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
Initial index, specified as a positive integer scalar. InitialIndex indicates the index of the first element from which the framing operation starts. Equivalently, InitialIndex-1 is the number of initial data samples in x to discard.
Incomplete frame rule, specified as either "drop" or
"padwithmissing". IncompleteFrameRule
specifies the rule to handle incomplete frames when the input does not have enough
remaining samples to fill up the last frame. If you set
IncompleteFrameRule to:
"drop"—framelbldrops the incomplete frame."padwithmissing"—framelblpads the last frame with missing values. Missing values are any of the following:<undefined>— For label values of typecategorical.<missing>— For label values of typestring.<false>— For label values of typelogical.<NaN>— For label values of a numeric type.
Data Types: char | string
Output Arguments
Framed label sequence, returned as a vector or a matrix.
Final condition array, returned as a vector.
If the overlap length is specified, then
fnlcondcontains the lastOverlapLengthsamples of the last complete frame and any samples that might remain from the incomplete frame, if there is one.If the underlap length is specified, then
fnlcondcontains only the remaining samples in the incomplete frame, if there are any. The final condition array has the same dimensions as the input label sequence. The final condition output can be used as the initial condition input for a subsequent framing operation in a sequence of consecutive framing operations. This allows the desired overlap to be maintained from one frame to the next.When
IncompleteFrameRuleis set to"padwithmissing", orInputLabelModeis set to"attibute", thenfnlcondis empty.
Final index output, returned as a positive integer scalar.
If the overlap length
OverlapLengthis specified,fnlidxis1.When you set
IncompleteFrameRuleto"padwithmissing"orInputLabelModeto"attribute", thenfnlidxis 1.fnlidx-1indicates the difference between the total number of points to skip between frames and the number of points inxthat were available to be skipped after filling the last frame. You can use the final index output as the initial index for a subsequent framing operation, thus maintaining the desired underlap from frame to frame.
Tips
framelbl complements the framesig function
in supervised-learning problems in which a label sequence is paired with a data sequence.
While framesig is used
to partition the data sequence into overlapping frames, framelbl is
used to perform the same framing operation on the label sequence.
Version History
Introduced in R2024a
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)