predict
Predict labels using k-nearest neighbor classification model
Description
label = predict(mdl,X)X, based on the trained k-nearest
neighbor classification model mdl. See Predicted Class Label.
[
also returns:label,score,cost]
= predict(mdl,X)
A matrix of classification scores (
score) indicating the likelihood that a label comes from a particular class. For k-nearest neighbor, scores are posterior probabilities. See Posterior Probability.A matrix of expected classification cost (
cost). For each observation inX, the predicted class label corresponds to the minimum expected classification costs among all classes. See Expected Cost.
Examples
Create a k-nearest neighbor classifier for Fisher's iris data, where k = 5. Evaluate some model predictions on new data.
Load the Fisher iris data set.
load fisheriris
X = meas;
Y = species;Create a classifier for five nearest neighbors. Standardize the noncategorical predictor data.
mdl = fitcknn(X,Y,'NumNeighbors',5,'Standardize',1);
Predict the classifications for flowers with minimum, mean, and maximum characteristics.
Xnew = [min(X);mean(X);max(X)]; [label,score,cost] = predict(mdl,Xnew)
label = 3×1 cell
{'versicolor'}
{'versicolor'}
{'virginica' }
score = 3×3
0.4000 0.6000 0
0 1.0000 0
0 0 1.0000
cost = 3×3
0.6000 0.4000 1.0000
1.0000 0 1.0000
1.0000 1.0000 0
The second and third rows of the score and cost matrices have binary values, which means all five nearest neighbors of the mean and maximum flower measurements have identical classifications.
Train k-nearest neighbor classifiers for various k values, and compare the decision boundaries of the classifiers.
Load the fisheriris data set.
load fisheririsThe data set contains length and width measurements from the sepals and petals of three species of iris flowers. Remove the sepal lengths and widths, and all observed setosa irises.
inds = ~strcmp(species,'setosa');
X = meas(inds,3:4);
species = species(inds); Create a binary label variable y. The label is 1 for a virginica iris and 0 for versicolor.
y = strcmp(species,'virginica');Train the k-nearest neighbor classifier. Specify 5 as the number of nearest neighbors to find, and standardize the predictor data.
EstMdl = fitcknn(X,y,'NumNeighbors',5,'Standardize',1)
EstMdl =
ClassificationKNN
ResponseName: 'Y'
CategoricalPredictors: []
ClassNames: [0 1]
ScoreTransform: 'none'
NumObservations: 100
Distance: 'euclidean'
NumNeighbors: 5
Properties, Methods
EstMdl is a trained ClassificationKNN classifier. Some of its properties appear in the Command Window.
Plot the decision boundary, which is the line that distinguishes between the two iris species based on their features.
x1 = min(X(:,1)):0.01:max(X(:,1)); x2 = min(X(:,2)):0.01:max(X(:,2)); [x1G,x2G] = meshgrid(x1,x2); XGrid = [x1G(:),x2G(:)]; pred = predict(EstMdl,XGrid); figure gscatter(XGrid(:,1),XGrid(:,2),pred,[1,0,0;0,0.5,1]) hold on plot(X(y == 0,1),X(y == 0,2),'ko', ... X(y == 1,1),X(y == 1,2),'kx') xlabel('Petal length (cm)') ylabel('Petal width (cm)') title('{\bf 5-Nearest Neighbor Classifier Decision Boundary}') legend('Versicolor Region','Virginica Region', ... 'Sampled Versicolor','Sampled Virginica', ... 'Location','best') axis tight hold off

The partition between the red and blue regions is the decision boundary. If you change the number of neighbors k, then the boundary changes.
Retrain the classifier using k = 1 (default value for NumNeighbors of fitcknn) and k = 20.
EstMdl1 = fitcknn(X,y); pred1 = predict(EstMdl1,XGrid); EstMdl20 = fitcknn(X,y,'NumNeighbors',20); pred20 = predict(EstMdl20,XGrid); figure gscatter(XGrid(:,1),XGrid(:,2),pred1,[1,0,0;0,0.5,1]) hold on plot(X(y == 0,1),X(y == 0,2),'ko', ... X(y == 1,1),X(y == 1,2),'kx') xlabel('Petal length (cm)') ylabel('Petal width (cm)') title('{\bf 1-Nearest Neighbor Classifier Decision Boundary}') legend('Versicolor Region','Virginica Region', ... 'Sampled Versicolor','Sampled Virginica', ... 'Location','best') axis tight hold off

figure gscatter(XGrid(:,1),XGrid(:,2),pred20,[1,0,0;0,0.5,1]) hold on plot(X(y == 0,1),X(y == 0,2),'ko', ... X(y == 1,1),X(y == 1,2),'kx') xlabel('Petal length (cm)') ylabel('Petal width (cm)') title('{\bf 20-Nearest Neighbor Classifier Decision Boundary}') legend('Versicolor Region','Virginica Region', ... 'Sampled Versicolor','Sampled Virginica', ... 'Location','best') axis tight hold off
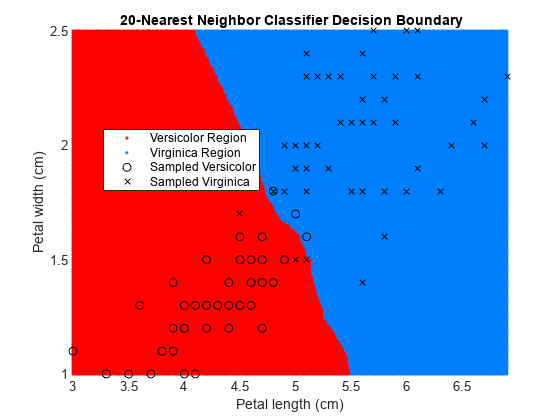
The decision boundary seems to linearize as k increases. This linearization happens because the algorithm down-weights the importance of each input with increasing k. When k = 1, the algorithm correctly predicts the species of almost all training samples. When k = 20, the algorithm has a higher misclassification rate within the training set. You can find an optimal value of k by using the OptimizeHyperparameters name-value argument of fitcknn. For an example, see Optimize Fitted KNN Classifier.
Input Arguments
k-nearest neighbor classifier model, specified as a
ClassificationKNN object.
Predictor data to be classified, specified as a numeric matrix or table.
Each row of X corresponds to one observation, and
each column corresponds to one variable.
For a numeric matrix:
The variables that make up the columns of
Xmust have the same order as the predictor variables used to trainmdl.If you train
mdlusing a table (for example,Tbl), thenXcan be a numeric matrix ifTblcontains all numeric predictor variables. k-nearest neighbor classification requires homogeneous predictors. Therefore, to treat all numeric predictors inTblas categorical during training, set'CategoricalPredictors','all'when you train usingfitcknn. IfTblcontains heterogeneous predictors (for example, numeric and categorical data types) andXis a numeric matrix, thenpredictthrows an error.
For a table:
predictdoes not support multicolumn variables and cell arrays other than cell arrays of character vectors.If you train
mdlusing a table (for example,Tbl), then all predictor variables inXmust have the same variable names and data types as those used to trainmdl(stored inmdl.PredictorNames). However, the column order ofXdoes not need to correspond to the column order ofTbl. BothTblandXcan contain additional variables (response variables, observation weights, and so on), butpredictignores them.If you train
mdlusing a numeric matrix, then the predictor names inmdl.PredictorNamesand corresponding predictor variable names inXmust be the same. To specify predictor names during training, see thePredictorNamesname-value pair argument offitcknn. All predictor variables inXmust be numeric vectors.Xcan contain additional variables (response variables, observation weights, and so on), butpredictignores them.
If you set 'Standardize',true in
fitcknn to train mdl, then the
software standardizes the columns of X using the
corresponding means in mdl.Mu and standard deviations in
mdl.Sigma.
Data Types: double | single | table
Output Arguments
Predicted class labels for the observations (rows) in
X, returned as a categorical array, character
array, logical vector, vector of numeric values, or cell array of character
vectors. label has length equal to the number of rows
in X.
For each observation, the label is the class with minimal expected cost.
For an observation with NaN scores, the
function classifies the observation into the majority class, which makes up the largest
proportion of the training labels.
Predicted class scores or posterior probabilities, returned as a numeric
matrix of size n-by-K.
n is the number of observations (rows) in
X, and K is the number of
classes (in mdl.ClassNames).
score(i,j) is the posterior probability that
observation i in X is of class
j in mdl.ClassNames. See Posterior Probability.
Data Types: single | double
Expected classification costs, returned as a numeric matrix of size
n-by-K. n is
the number of observations (rows) in X, and
K is the number of classes (in
mdl.ClassNames). cost(i,j) is the
cost of classifying row i of X as
class j in mdl.ClassNames. See Expected Cost.
Data Types: single | double
Algorithms
predict classifies by minimizing the expected
misclassification cost:
where:
is the predicted classification.
K is the number of classes.
is the posterior probability of class j for observation x.
is the cost of classifying an observation as y when its true class is j.
Consider a vector (single query point) xnew and a model
mdl.
k is the number of nearest neighbors used in prediction,
mdl.NumNeighbors.nbd(mdl,xnew)specifies the k nearest neighbors toxnewinmdl.X.Y(nbd)specifies the classifications of the points innbd(mdl,xnew), namelymdl.Y(nbd).W(nbd)specifies the weights of the points innbd(mdl,xnew).priorspecifies the priors of the classes inmdl.Y.
If the model contains a vector of prior probabilities, then the observation weights
W are normalized by class to sum to the priors.
This process might involve a calculation for the point xnew,
because weights can depend on the distance from xnew to the
points in mdl.X.
The posterior probability p(j|xnew)
is
Here, is 1 when
mdl.Y(i) = j, and
0 otherwise.
Two costs are associated with KNN classification: the true misclassification cost per class and the expected misclassification cost per observation.
You can set the true misclassification cost per class by using the 'Cost'
name-value pair argument when you run fitcknn. The value Cost(i,j) is the cost of classifying
an observation into class j if its true class is i. By
default, Cost(i,j) = 1 if i ~= j, and
Cost(i,j) = 0 if i = j. In other words, the cost
is 0 for correct classification and 1 for incorrect
classification.
Two costs are associated with KNN classification: the true misclassification cost per class
and the expected misclassification cost per observation. The third output of predict is the expected misclassification cost per
observation.
Suppose you have Nobs observations that you want to classify with a trained
classifier mdl, and you have K classes. You place the
observations into a matrix Xnew with one observation per row. The
command
[label,score,cost] = predict(mdl,Xnew)
returns a matrix cost of size
Nobs-by-K, among other outputs. Each row of the
cost matrix contains the expected (average) cost of classifying the
observation into each of the K classes. cost(n,j)
is
where
K is the number of classes.
is the posterior probability of class i for observation Xnew(n).
is the true misclassification cost of classifying an observation as j when its true class is i.
Alternative Functionality
Simulink Block
To integrate the prediction of a nearest neighbor classification model into
Simulink®, you can use the ClassificationKNN
Predict block in the Statistics and Machine Learning Toolbox™ library or a MATLAB® Function block with the predict function. For
examples, see Predict Class Labels Using ClassificationKNN Predict Block and Predict Class Labels Using MATLAB Function Block.
When deciding which approach to use, consider the following:
If you use the Statistics and Machine Learning Toolbox library block, you can use the Fixed-Point Tool (Fixed-Point Designer) to convert a floating-point model to fixed point.
Support for variable-size arrays must be enabled for a MATLAB Function block with the
predictfunction.If you use a MATLAB Function block, you can use MATLAB functions for preprocessing or post-processing before or after predictions in the same MATLAB Function block.
Extended Capabilities
The
predict function fully supports tall arrays. For more information,
see Tall Arrays.
Usage notes and limitations:
Use
saveLearnerForCoder,loadLearnerForCoder, andcodegen(MATLAB Coder) to generate code for thepredictfunction. Save a trained model by usingsaveLearnerForCoder. Define an entry-point function that loads the saved model by usingloadLearnerForCoderand calls thepredictfunction. Then usecodegento generate code for the entry-point function.To generate single-precision C/C++ code for
predict, specifyDataType="single"when you call theloadLearnerForCoderfunction.This table contains notes about the arguments of
predict. Arguments not included in this table are fully supported.Argument Notes and Limitations mdlA
ClassificationKNNmodel object is a full object that does not have a corresponding compact object. For this model,saveLearnerForCodersaves a compact version that does not include the hyperparameter optimization properties.If
mdlis a model trained using the kd-tree search algorithm, and the code generation build type is a MEX function, thencodegen(MATLAB Coder) generates a MEX function using Intel® Threading Building Blocks (TBB) for parallel computation. Otherwise,codegengenerates code usingparfor(MATLAB Coder).MEX function for the kd-tree search algorithm —
codegengenerates an optimized MEX function using Intel TBB for parallel computation on multicore platforms. You can use the MEX function to accelerate MATLAB algorithms. For details on Intel TBB, see https://www.intel.com/content/www/us/en/developer/tools/oneapi/onetbb.html.If you generate the MEX function to test the generated code of the
parforversion, you can disable the usage of Intel TBB. Set theExtrinsicCallsproperty of the MEX configuration object tofalse. For details, seecoder.MexCodeConfig(MATLAB Coder).MEX function for the exhaustive search algorithm and standalone C/C++ code for both algorithms — The generated code of
predictusesparfor(MATLAB Coder) to create loops that run in parallel on supported shared-memory multicore platforms in the generated code. If your compiler does not support the Open Multiprocessing (OpenMP) application interface or you disable OpenMP library, MATLAB Coder™ treats theparfor-loops asfor-loops. To find supported compilers, see Supported Compilers. To disable OpenMP library, set theEnableOpenMPproperty of the configuration object tofalse. For details, seecoder.CodeConfig(MATLAB Coder).
For the usage notes and limitations of the model object, see Code Generation of the
ClassificationKNNobject.
XXmust be a single-precision or double-precision matrix or a table containing numeric variables, categorical variables, or both.The number of rows, or observations, in
Xcan be a variable size, but the number of columns inXmust be fixed.If you want to specify
Xas a table, then your model must be trained using a table, and your entry-point function for prediction must do the following:Accept data as arrays.
Create a table from the data input arguments and specify the variable names in the table.
Pass the table to
predict.
For an example of this table workflow, see Generate Code to Classify Data in Table. For more information on using tables in code generation, see Code Generation for Tables (MATLAB Coder) and Table Limitations for Code Generation (MATLAB Coder).
For more information, see Introduction to Code Generation.
Usage notes and limitations:
predictdoes not support GPU arrays forClassificationKNNmodels with the following specifications:The
NSMethodproperty is specified as"kdtree".The
Distanceproperty is specified as"fasteuclidean","fastseuclidean", or a function handle.The
IncludeTiesproperty is specified astrue.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2012a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)