loss
Classification loss for classification tree model
Syntax
Description
L = loss(tree,Tbl,ResponseVarName)L for the trained classification tree model
tree using the predictor data in table
Tbl and the true class labels in
Tbl.ResponseVarName. The interpretation of
L depends on the loss function
(LossFun) and weighting scheme
(Weights). In general, better classifiers yield smaller
classification loss values.
L = loss(___,Name=Value)
Examples
Compute the resubstituted classification error for the ionosphere data set.
load ionosphere
tree = fitctree(X,Y);
L = loss(tree,X,Y)L = 0.0114
Unpruned decision trees tend to overfit. One way to balance model complexity and out-of-sample performance is to prune a tree (or restrict its growth) so that in-sample and out-of-sample performance are satisfactory.
Load Fisher's iris data set. Partition the data into training (50%) and validation (50%) sets.
load fisheriris n = size(meas,1); rng(1) % For reproducibility idxTrn = false(n,1); idxTrn(randsample(n,round(0.5*n))) = true; % Training set logical indices idxVal = idxTrn == false; % Validation set logical indices
Grow a classification tree using the training set.
Mdl = fitctree(meas(idxTrn,:),species(idxTrn));
View the classification tree.
view(Mdl,'Mode','graph');

The classification tree has four pruning levels. Level 0 is the full, unpruned tree (as displayed). Level 3 is just the root node (i.e., no splits).
Examine the training sample classification error for each subtree (or pruning level) excluding the highest level.
m = max(Mdl.PruneList) - 1;
trnLoss = resubLoss(Mdl,'Subtrees',0:m)trnLoss = 3×1
0.0267
0.0533
0.3067
The full, unpruned tree misclassifies about 2.7% of the training observations.
The tree pruned to level 1 misclassifies about 5.3% of the training observations.
The tree pruned to level 2 (i.e., a stump) misclassifies about 30.6% of the training observations.
Examine the validation sample classification error at each level excluding the highest level.
valLoss = loss(Mdl,meas(idxVal,:),species(idxVal),'Subtrees',0:m)valLoss = 3×1
0.0369
0.0237
0.3067
The full, unpruned tree misclassifies about 3.7% of the validation observations.
The tree pruned to level 1 misclassifies about 2.4% of the validation observations.
The tree pruned to level 2 (i.e., a stump) misclassifies about 30.7% of the validation observations.
To balance model complexity and out-of-sample performance, consider pruning Mdl to level 1.
pruneMdl = prune(Mdl,'Level',1); view(pruneMdl,'Mode','graph')

Input Arguments
Trained classification tree, specified as a ClassificationTree model object trained with fitctree, or a CompactClassificationTree model object
created with compact.
Sample data, specified as a table. Each row of Tbl corresponds to
one observation, and each column corresponds to one predictor variable. Optionally,
Tbl can contain additional columns for the response variable
and observation weights. Tbl must contain all the predictors used
to train tree. Multicolumn variables and cell arrays other than
cell arrays of character vectors are not allowed.
If Tbl contains the response variable used to train
tree, then you do not need to specify
ResponseVarName or Y.
If you train tree using sample data contained in a table, then
the input data for loss must also be in a table.
Data Types: table
Response variable name, specified as the name of a variable in Tbl. If
Tbl contains the response variable used to train
tree, then you do not need to specify
ResponseVarName.
You must specify ResponseVarName as a character vector or string scalar.
For example, if the response variable is stored as Tbl.Response, then
specify it as "Response". Otherwise, the software treats all columns
of Tbl, including Tbl.Response, as
predictors.
The response variable must be a categorical, character, or string array, a logical or numeric vector, or a cell array of character vectors. If the response variable is a character array, then each element must correspond to one row of the array.
Data Types: char | string
Class labels, specified as a categorical, character, or string array, a logical or numeric
vector, or a cell array of character vectors. Y must be
of the same type as the class labels used to train
tree, and its number of elements must equal the number
of rows of X.
Data Types: categorical | char | string | logical | single | double | cell
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: L = loss(tree,X,Y,LossFun="exponential") specifies to
use an exponential loss function.
Loss function, specified as a built-in loss function name or a function handle.
The following table describes the values for the built-in loss functions.
| Value | Description |
|---|---|
"binodeviance" | Binomial deviance |
"classifcost" | Observed misclassification cost |
"classiferror" | Misclassified rate in decimal |
"exponential" | Exponential loss |
"hinge" | Hinge loss |
"logit" | Logistic loss |
"mincost" | Minimal expected misclassification cost (for classification scores that are posterior probabilities) |
"quadratic" | Quadratic loss |
"mincost" is appropriate for
classification scores that are posterior probabilities. Classification
trees return posterior probabilities as classification scores by default
(see predict).
Specify your own function using function handle notation. Suppose that
n is the number of observations in
X, and K is the number of
distinct classes (numel(tree.ClassNames)). Your
function must have the signature
lossvalue = lossfun(C,S,W,Cost)The output argument
lossvalueis a scalar.You specify the function name (
lossfun).Cis an n-by-K logical matrix with rows indicating the class to which the corresponding observation belongs. The column order corresponds to the class order intree.ClassNames.Create
Cby settingC(p,q) = 1, if observationpis in classq, for each row. Set all other elements of rowpto0.Sis an n-by-K numeric matrix of classification scores. The column order corresponds to the class order intree.ClassNames.Sis a matrix of classification scores, similar to the output ofpredict.Wis an n-by-1 numeric vector of observation weights. If you passW, the software normalizes the weights to sum to1.Costis a K-by-K numeric matrix of misclassification costs. For example,Cost = ones(K) - eye(K)specifies a cost of0for correct classification and1for misclassification.
For more details on the loss functions, see Classification Loss.
Example: LossFun="binodeviance"
Example: LossFun=@lossfun
Data Types: char | string | function_handle
Observation weights, specified as a numeric vector or the name of a variable in Tbl.
If you specify Weights as a numeric vector, then the size of Weights must be equal to the number of rows in X or Tbl.
If you specify Weights as the name of a variable in
Tbl, then the name must be a character vector or string scalar.
For example, if the weights are stored as Tbl.W, then specify it as
"W". Otherwise, the software treats all columns of
Tbl, including Tbl.W, as
predictors.
loss normalizes the weights so that the observation weights in
each class sum to the prior probability of that class. When you specify
Weights, loss computes the weighted
classification loss.
Example: Weights="W"
Data Types: single | double | char | string
Pruning level, specified as a vector of nonnegative integers in ascending order or
"all".
If you specify a vector, then all elements must be at least 0 and
at most max(tree.PruneList). 0 indicates the full,
unpruned tree, and max(tree.PruneList) indicates the completely
pruned tree (that is, just the root node).
If you specify "all", then loss
operates on all subtrees (in other words, the entire pruning sequence). This
specification is equivalent to using 0:max(tree.PruneList).
loss prunes tree to each level
specified by Subtrees, and then estimates the corresponding output
arguments. The size of Subtrees determines the size of some output
arguments.
For the function to invoke Subtrees, the properties
PruneList and PruneAlpha of
tree must be nonempty. In other words, grow
tree by setting Prune="on" when you use
fitctree, or by pruning tree using prune.
Example: Subtrees="all"
Data Types: single | double | char | string
Tree size, specified as one of these values:
"se"—lossreturns the best pruning level (BestLevel), which corresponds to the highest pruning level with the loss within one standard deviation of the minimum (L+se, whereLandserelate to the smallest value inSubtrees)."min"—lossreturns the best pruning level, which corresponds to the element ofSubtreeswith the smallest loss. This element is usually the smallest element ofSubtrees.
Example: TreeSize="min"
Data Types: char | string
Output Arguments
Classification
loss, returned as a numeric vector that has the same length as
Subtrees. The meaning of the error depends on the
values in Weights and
LossFun.
Standard error of loss, returned as a numeric vector that has the same
length as Subtrees.
Number of leaf nodes in the pruned subtrees, returned as a vector of
integer values that has the same length as Subtrees.
Leaf nodes are terminal nodes, which give responses, not splits.
Best pruning level, returned as a numeric scalar whose value depends on
TreeSize:
When
TreeSizeis"se", thelossfunction returns the highest pruning level whose loss is within one standard deviation of the minimum (L+se, whereLandserelate to the smallest value inSubtrees).When
TreeSizeis"min", thelossfunction returns the element ofSubtreeswith the smallest loss, usually the smallest element ofSubtrees.
More About
Classification loss functions measure the predictive inaccuracy of classification models. When you compare the same type of loss among many models, a lower loss indicates a better predictive model.
Consider the following scenario.
L is the weighted average classification loss.
n is the sample size.
For binary classification:
yj is the observed class label. The software codes it as –1 or 1, indicating the negative or positive class (or the first or second class in the
ClassNamesproperty), respectively.f(Xj) is the positive-class classification score for observation (row) j of the predictor data X.
mj = yjf(Xj) is the classification score for classifying observation j into the class corresponding to yj. Positive values of mj indicate correct classification and do not contribute much to the average loss. Negative values of mj indicate incorrect classification and contribute significantly to the average loss.
For algorithms that support multiclass classification (that is, K ≥ 3):
yj* is a vector of K – 1 zeros, with 1 in the position corresponding to the true, observed class yj. For example, if the true class of the second observation is the third class and K = 4, then y2* = [
0 0 1 0]′. The order of the classes corresponds to the order in theClassNamesproperty of the input model.f(Xj) is the length K vector of class scores for observation j of the predictor data X. The order of the scores corresponds to the order of the classes in the
ClassNamesproperty of the input model.mj = yj*′f(Xj). Therefore, mj is the scalar classification score that the model predicts for the true, observed class.
The weight for observation j is wj. The software normalizes the observation weights so that they sum to the corresponding prior class probability stored in the
Priorproperty. Therefore,
Given this scenario, the following table describes the supported loss functions that you can specify by using the LossFun name-value argument.
| Loss Function | Value of LossFun | Equation |
|---|---|---|
| Binomial deviance | "binodeviance" | |
| Observed misclassification cost | "classifcost" | where is the class label corresponding to the class with the maximal score, and is the user-specified cost of classifying an observation into class when its true class is yj. |
| Misclassified rate in decimal | "classiferror" | where I{·} is the indicator function. |
| Cross-entropy loss | "crossentropy" |
The weighted cross-entropy loss is where the weights are normalized to sum to n instead of 1. |
| Exponential loss | "exponential" | |
| Hinge loss | "hinge" | |
| Logistic loss | "logit" | |
| Minimal expected misclassification cost | "mincost" |
The software computes the weighted minimal expected classification cost using this procedure for observations j = 1,...,n.
The weighted average of the minimal expected misclassification cost loss is |
| Quadratic loss | "quadratic" |
If you use the default cost matrix (whose element value is 0 for correct classification
and 1 for incorrect classification), then the loss values for
"classifcost", "classiferror", and
"mincost" are identical. For a model with a nondefault cost matrix,
the "classifcost" loss is equivalent to the "mincost"
loss most of the time. These losses can be different if prediction into the class with
maximal posterior probability is different from prediction into the class with minimal
expected cost. Note that "mincost" is appropriate only if classification
scores are posterior probabilities.
This figure compares the loss functions (except "classifcost",
"crossentropy", and "mincost") over the score
m for one observation. Some functions are normalized to pass through
the point (0,1).
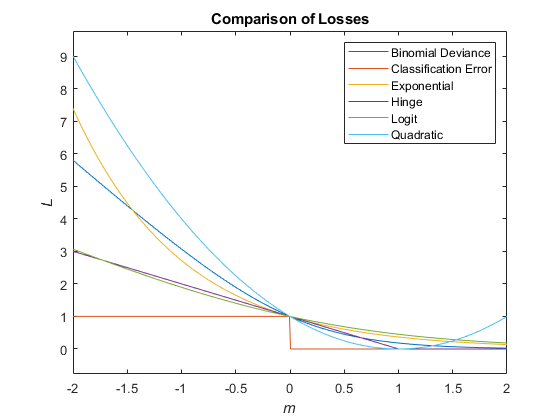
The true misclassification cost is the cost of classifying an observation into an incorrect class.
You can set the true misclassification cost per class by using the Cost
name-value argument when you create the classifier. Cost(i,j) is the cost
of classifying an observation into class j when its true class is
i. By default, Cost(i,j)=1 if
i~=j, and Cost(i,j)=0 if i=j.
In other words, the cost is 0 for correct classification and
1 for incorrect classification.
The expected misclassification cost per observation is an averaged cost of classifying the observation into each class.
Suppose you have Nobs observations that you want to classify with a trained
classifier, and you have K classes. You place the observations
into a matrix X with one observation per row.
The expected cost matrix CE has size
Nobs-by-K. Each row of
CE contains the expected (average) cost of classifying
the observation into each of the K classes.
CE(n,k)
is
where:
K is the number of classes.
is the posterior probability of class i for observation X(n).
is the true misclassification cost of classifying an observation as k when its true class is i.
For trees, the score of a classification of a leaf node is the posterior probability of the classification at that node. The posterior probability of the classification at a node is the number of training sequences that lead to that node with the classification, divided by the number of training sequences that lead to that node.
For an example, see Posterior Probability Definition for Classification Tree.
Extended Capabilities
The
loss function supports tall arrays with the following usage
notes and limitations:
Only one output is supported.
You can use models trained on either in-memory or tall data with this function.
For more information, see Tall Arrays.
Usage notes and limitations:
The
lossfunction does not support decision tree models trained with surrogate splits.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2011a
See Also
margin | edge | predict | fitctree | ClassificationTree | CompactClassificationTree
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)