Extract Answers from Documents Using BERT
This example shows how to modify and fine-tune a pretrained BERT model for extractive question answering. In extractive question answering, you provide a model with a question and a source document that contains the answer. Then, the model finds the exact extract of the source document that answers the question.
The dataset used in this example is the Stanford Question Answering Dataset (SQuAD) 2.0 dataset [1].
Import Data
Load the preprocessed SQuAD 2.0 data.
dataFolder = fullfile(tempdir,"squad2_data"); if ~datasetExists(dataFolder) zipFile = matlab.internal.examples.downloadSupportFile("textanalytics","data/squad2_data.zip"); unzip(zipFile,dataFolder); end data = load(fullfile(dataFolder,"squad2_data.mat"));
The preprocessed dataset includes training and validation data.
trainData = data.trainingData; validationData = data.valData;
Print the first question in the validation dataset.
validationData.Question(1)
ans = "In what country is Normandy located?"
Print the first context in the validation dataset. This information contains the answer to the question, starting from the AnswerStart index.
validationData.Context(1)
ans =
"The Normans (Norman: Nourmands; French: Normands; Latin: Normanni) were the people who in the 10th and 11th centuries gave their name to Normandy, a region in France. They were descended from Norse ("Norman" comes from "Norseman") raiders and pirates from Denmark, Iceland and Norway who, under their leader Rollo, agreed to swear fealty to King Charles III of West Francia. Through generations of assimilation and mixing with the native Frankish and Roman-Gaulish populations, their descendants would gradually merge with the Carolingian-based cultures of West Francia. The distinct cultural and ethnic identity of the Normans emerged initially in the first half of the 10th century, and it continued to evolve over the succeeding centuries."
validationData.AnswerStart(1)
ans = 159
Print the answer to the question.
validationData.Answer(1)
ans = "France"
To evaluate the model after training, split the validation data into a smaller validation set and a test set.
cv = cvpartition(length(validationData.Question),Holdout=0.2); idxTest = cv.test; testData = validationData(idxTest,:); validationData = validationData(~idxTest,:);
Load and Modify Pretrained BERT Model
Load a pretrained BERT-tiny model using the bert function. If the Text Analytics Toolbox™ Model for BERT-Tiny Network support package is not installed, then the function provides a download link.
[net,tokenizer] = bert(Model="tiny");In extractive question answering, you predict the start and end indices of the answer within the context. To do this, add a fully connected layer with two outputs to the end of your network.
outputLayer = net.OutputNames{1};
net = addLayers(net,fullyConnectedLayer(2));
net = connectLayers(net,outputLayer,"fc");
net = initialize(net);Prepare Data for Training
Tokenize and preprocess the data using the helper function prepareData, defined at the end of this example. This process can take a few minutes.
[trainInputID,trainSegmentID,trainStartIdx,trainEndIdx] = prepareData(trainData,tokenizer); [validationInputID,validationSegmentID,validationStartIdx,validationEndIdx] = prepareData(validationData,tokenizer); [testInputID,testSegmentID,testStartIdx,testEndIdx] = prepareData(testData,tokenizer);
Create BERT Mini-Batch Queue
Create minibatchqueue objects for the training and validation data to feed into the model during training.
mbqTrain = bertMiniBatchQueueForTraining(trainInputID,trainSegmentID,trainStartIdx,trainEndIdx,tokenizer.PaddingCode); mbqValidation = bertMiniBatchQueueForTraining(validationInputID,validationSegmentID,validationStartIdx,validationEndIdx,tokenizer.PaddingCode);
Specify Training Options
Specify the training options. Choosing among the options requires empirical analysis. To explore different training option configurations by running experiments, you can use the Experiment Manager app.
Train using the Adam optimizer.
Train for five epochs.
For fine-tuning, lower the learning rate. Train using a learning rate of
2e-5.Set the mini-batch size to
32.Validate the network using the validation data.
Shuffle the data every epoch.
Monitor the training progress in a plot and monitor the custom metric
exactMatchfor evaluation. This function is defined at the end of this example.Disable the verbose output.
miniBatchSize = 32; numEpochs = 5; learnRate = 2e-5; options = trainingOptions("adam", ... MaxEpochs=numEpochs, ... InitialLearnRate=learnRate, ... MiniBatchSize=miniBatchSize, ... ValidationData=mbqValidation, ... Shuffle="every-epoch", ... Metrics=@exactMatch, ... Plots="training-progress", ... Verbose=false);
Define the custom loss function. The questionAnsweringLoss function computes the loss for question answering tasks by comparing the predicted start and end positions of answers with the true positions using a cross-entropy loss function.
function L = questionAnsweringLoss(Y,T) YStart = dlarray(stripdims(reshape(Y(1,:,:),[],size(Y,3))),"BC"); YEnd = dlarray(stripdims(reshape(Y(2,:,:),[],size(Y,3))),"BC"); TStart = T(1,:); TEnd = T(2,:); YStart = softmax(YStart); YEnd = softmax(YEnd); LStart = indexcrossentropy(YStart,TStart); LEnd = indexcrossentropy(YEnd,TEnd); L = (LStart + LEnd) / 2; end
Train Network
Train the BERT network.
net = trainnet(mbqTrain,net,@(Y,T) questionAnsweringLoss(Y,T),options);
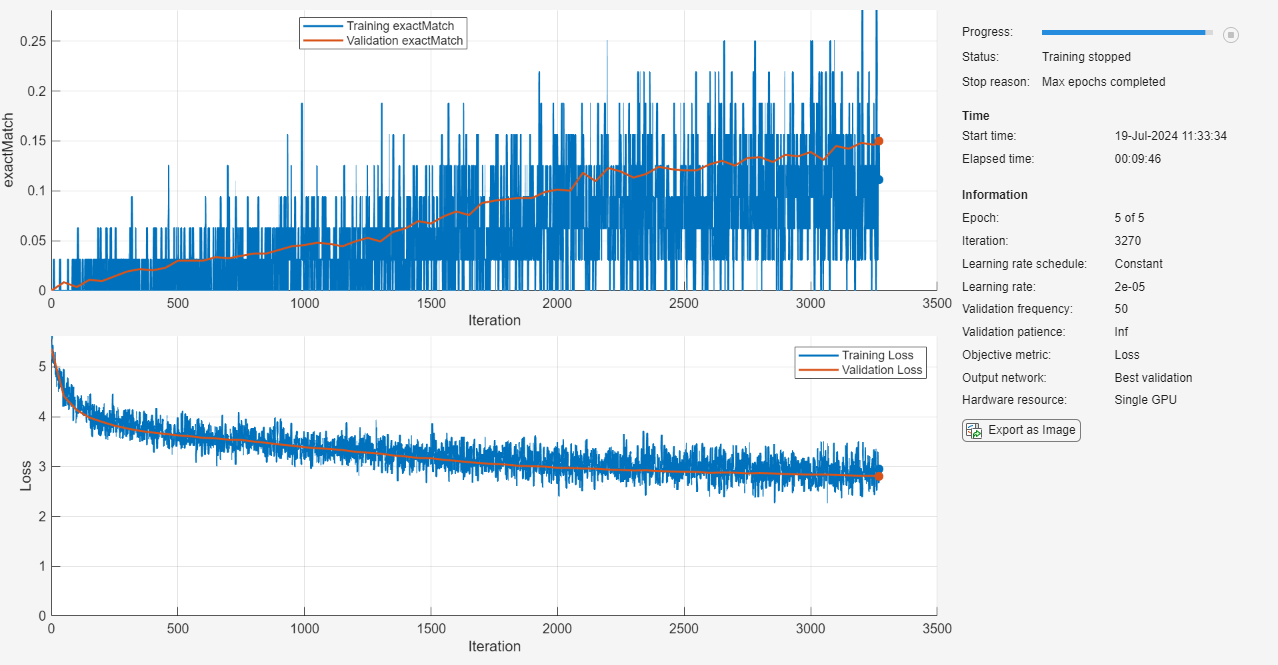
Test Network
Create a mini-batch queue suitable for prediction from the input data.
mbqTest = bertMiniBatchQueueForPrediction(testInputID,testSegmentID,tokenizer)
mbqTest =
minibatchqueue with 3 outputs and properties:
Mini-batch creation:
MiniBatchSize: 128
PartialMiniBatch: 'return'
MiniBatchFcn: @(inputIds,segmentIds)preprocessPredictors(inputIds,segmentIds,paddingValue)
PreprocessingEnvironment: 'serial'
Outputs:
OutputCast: {'single' 'single' 'single'}
OutputAsDlarray: [1 1 1]
MiniBatchFormat: {'CTB' 'CTB' 'CTB'}
OutputEnvironment: {'auto' 'auto' 'auto'}
Evaluate the model on the test data and calculate the exact match metric.
YPred = minibatchpredict(net,mbqTest,MiniBatchSize=miniBatchSize,UniformOutput=false); YTest = [testStartIdx testEndIdx];
Compare all the batches and combine the results to obtain the average results across all batches.
totalExactMatch = 0; for k=1:length(YPred) totalExactMatch = totalExactMatch + exactMatch(YPred{k}, YTest(k,:)); end testExactMatch = totalExactMatch/length(YPred)
testExactMatch =
1(C) × 1(B) × 1(T) dlarray
0.1528
Answer Question
You can use the fine-tuned model to predict the answers to new questions.
question = wordTokenize(tokenizer,"When was the Hubble Space Telescope launched?"); context = wordTokenize(tokenizer,"The Hubble Space Telescope is a large telescope in space launched in 1990." + ... " It is one of the largest and most versatile telescopes, renowned for its deep space images" + ... " and has made many astronomical discoveries.");
Predict the start and end tokens.
[inputIds, segIds, idx2words] = encodeTokens(tokenizer,question,context); attMask = ones("like",segIds{1}); Y = predict(net,inputIds{1},segIds{1},attMask,InputDataFormats=["CTB","CTB","CTB"]); [~, idx] = max(Y, [], 2); YStart = idx(1)
YStart = 25
YEnd = idx(2)
YEnd = 25
Convert the obtained tokens back to the original words and display the answer.
idx2words = idx2words{1};
answerIdx = unique(idx2words(YStart:YEnd));
context = context{1};
answer = context(answerIdx)answer = "1990"
Helper Functions
The prepareData function tokenizes the questions and contexts from the SQuAD dataset using the input tokenizer and finds the token-level positions of the answers. For this example, use short contexts with a maximum of 128 tokens.
function [allInputIds, allSegIds, allStartIdx, allEndIdx] = prepareData(data, tokenizer) maxSeqLength = 128; numElements = height(data); allInputIds = cell(numElements,1); allSegIds = cell(numElements,1); allStartIdx = zeros(numElements,1); allEndIdx = zeros(numElements,1); % Process each data element for i = 1:numElements question = data(i,:).Question; context = data(i,:).Context; answer = data(i,:).Answer; % Tokenize and truncate input [inputIds, segIds] = encode(tokenizer,question,context); inputIds = inputIds{:}; segIds = segIds{:}; % Only process if within max sequence length if numel(inputIds) <= maxSeqLength tokenizedAnswer = encode(tokenizer,answer,AddSpecialTokens=false); tokenizedAnswer = tokenizedAnswer{:}; indices = strfind(inputIds,tokenizedAnswer); % If answer is found in the context if ~isempty(indices) startIndex = indices(1); endIndex = startIndex + numel(tokenizedAnswer) - 1; allInputIds{i} = inputIds; allSegIds{i} = segIds; allStartIdx(i) = startIndex; allEndIdx(i) = endIndex; end end end % Remove empty entries validEntries = allStartIdx > 0; allInputIds = allInputIds(validEntries); allSegIds = allSegIds(validEntries); allStartIdx = allStartIdx(validEntries); allEndIdx = allEndIdx(validEntries); end
The exactMatch function computes the exact match metric, which measures whether the predicted start and end positions of answers match the true positions exactly.
function val = exactMatch(Y,T) batchSize = size(Y,finddim(Y,"B")); [~, idx] = max(Y,[],3); val = idx == T; val = all(val,1); val = sum(val)/batchSize; end
The bertMiniBatchQueueForTraining function creates a minibatchqueue object from the documents and labels to train a BERT model for question answering. The bertMiniBatchQueueForPrediction function creates a minibatchqueue object from the documents to make predictions with a trained BERT model.
function mbq = bertMiniBatchQueueForTraining(inputIDs,segmentIDs,startIdx,endIdx,paddingValue) inputIDsDS = arrayDatastore(inputIDs,OutputType="same"); segIDsDS = arrayDatastore(segmentIDs,OutputType="same"); Y = arrayDatastore([startIdx endIdx]); cds = combine(inputIDsDS,segIDsDS,Y); mbq = minibatchqueue(cds,4,... MiniBatchFcn=@(inputIds,segmentIds,targets) preprocessMiniBatch(inputIds, ... segmentIds, ... targets, ... paddingValue), ... MiniBatchFormat=["CTB" "CTB" "CTB" "BC"]); end function mbq = bertMiniBatchQueueForPrediction(inputIDs,segmentIDs,paddingValue) inputIDsDS = arrayDatastore(inputIDs,OutputType="same"); segIDsDS = arrayDatastore(segmentIDs,OutputType="same"); cds = combine(inputIDsDS,segIDsDS); mbq = minibatchqueue(cds,3,... MiniBatchFcn=@(inputIds,segmentIds) preprocessPredictors(inputIds, ... segmentIds, ... paddingValue), ... MiniBatchFormat=["CTB" "CTB" "CTB"]); end function [inputIDs, mask, segmentIDs, targets] = preprocessMiniBatch(inputIDs,segmentIDs,targets,paddingValue) [inputIDs, mask] = padsequences(inputIDs,2,PaddingValue=paddingValue); segmentIDs = padsequences(segmentIDs,2,PaddingValue=paddingValue); targets = cell2mat(targets); end
References
[1] Rajpurkar, Pranav, Robin Jia, and Percy Liang. "Know What You Don’t Know: Unanswerable Questions for SQuAD." Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers). Association for Computational Linguistics, 2018.
See Also
bert | trainnet (Deep Learning Toolbox) | wordTokenize | encodeTokens | trainingOptions (Deep Learning Toolbox) | minibatchqueue (Deep Learning Toolbox) | minibatchpredict (Deep Learning Toolbox)