msresample
Resample signal with peaks
Syntax
[Xout, Intensitiesout]
= msresample(X, Intensities, N)
msresample(..., 'Uniform', UniformValue,
...)
msresample(..., 'Range', RangeValue,
...)
msresample(..., 'RangeWarnOff', RangeWarnOffValue,
...)
msresample(..., 'Missing', MissingValue,
...)
msresample(..., 'Window', WindowValue,
...)
msresample(..., 'Cutoff', CutoffValue,
...)
msresample(..., 'ShowPlot', ShowPlotValue,
...)
Arguments
X | Vector of separation-unit values for
a set of signals with peaks. The number of elements in the vector
equals the number of rows in the matrix Intensities.
The separation unit can quantify wavelength, frequency, distance,
time, or m/z depending on the instrument that generates the signal
data. |
Intensities | Matrix of intensity values for a set
of peaks that share the same separation-unit range. Each row corresponds
to a separation-unit value, and each column corresponds to either
a set of signals with peaks or a retention time. The number of rows
equals the number of elements in vector X. |
N | Positive integer specifying the total number of samples. |
Description
Tip
Use the following syntaxes with data from any separation technique that produces signal data, such as spectroscopy, NMR, electrophoresis, chromatography, or mass spectrometry.
[ resamples
raw noisy signal data, Xout, Intensitiesout]
= msresample(X, Intensities, N)Intensities. The
output signal has N samples with a spacing
that increases linearly within the range [min(. X)
max(X)]X can
be a linear or a quadratic function of its index. When you set input
arguments such that down-sampling takes place, msresample applies
a lowpass filter before resampling to minimize aliasing.
For the antialias filter, msresample uses
a linear-phase FIR filter with a least-squares error minimization.
The cutoff frequency is set by the largest down-sampling ratio when
comparing the same regions in the X and
Xout vectors.
Tip
msresample is particularly useful when you
have signals with different separation-unit vectors and you want to
match the scales.
msresample(..., ' calls PropertyName', PropertyValue,
...)msresample with optional properties
that use property name/property value pairs. You can specify one or
more properties in any order. Each PropertyName must
be enclosed in single quotes and is case insensitive. These property
name/property value pairs are as follows:
msresample(..., 'Uniform', , when UniformValue,
...)UniformValuetrue,
it forces the vector X to be uniformly
spaced. The default value is false.
msresample(..., 'Range', specifies a RangeValue,
...)1-by-2 vector
with the separation-unit range for the output signal, Intensitiesout. RangeValue must
be within [min(].
Default value is the full range X) max(X)[min(. When X)
max(X)]RangeValue values
exceed the values in X, msresample extrapolates
the signal with zeros and returns a warning message.
msresample(..., 'RangeWarnOff', controls the return of a warning message when RangeWarnOffValue,
...)RangeValue values
exceed the values in X. RangeWarnOffValue can
be true or false (default).
msresample(..., 'Missing', , when MissingValue,
...)MissingValuetrue,
analyzes the input vector, X, for dropped
samples. The default value is false. If the down-sample
factor is large, checking for dropped samples might not be worth the
extra computing time. Dropped samples can only be recovered if the
original separation-unit values follow a linear or a quadratic function
of the X vector index.
msresample(..., 'Window', specifies the window used when calculating parameters
for the lowpass filter. Enter WindowValue,
...)'Flattop', 'Blackman', 'Hamming',
or 'Hanning'. The default value is 'Flattop'.
msresample(..., 'Cutoff', specifies the cutoff frequency. Enter a scalar value
from CutoffValue,
...)0 to 1 (Nyquist frequency
or half the sampling frequency). By default, msresample estimates
the cutoff value by inspecting the separation-unit vectors, X and XOut.
However, the cutoff frequency might be underestimated if X has
anomalies.
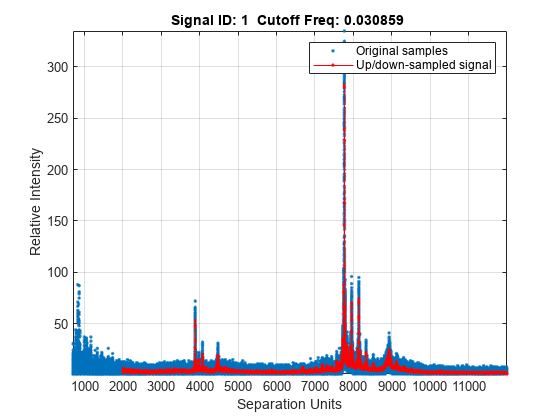
msresample(..., 'ShowPlot', plots the original and the resampled signal. When ShowPlotValue,
...)msresample is
called without output arguments, the signals are plotted unless ShowPlotValuefalse.
When ShowPlotValuetrue,
only the first signal in Intensities is
plotted. ShowPlotValueIntensities.
Tip
LC/MS data analysis requires extended amounts of memory from the operating system.
If you receive errors related to memory, try the following:
Increase the virtual memory (swap space) for your operating system as described in Resolve “Out of Memory” Errors.
If you receive errors related to Java® heap space, increase your Java heap space:
If you have MATLAB® version 7.10 (R2010a) or later, see Java Heap Memory Settings.
If you have MATLAB version 7.9 (R2009b) or earlier, see https://www.mathworks.com/matlabcentral/answers/92813-how-do-i-increase-the-heap-space-for-the-java-vm-in-matlab.
Examples
Version History
Introduced before R2006a
See Also
mspalign | msbackadj | msdotplot | msalign | msheatmap | mslowess | msnorm | mspeaks | msppresample | mssgolay | msviewer
Topics
- Mass Spectrometry and Bioanalytics
- Preprocessing Raw Mass Spectrometry Data
- Visualizing and Preprocessing Hyphenated Mass Spectrometry Data Sets for Metabolite and Protein/Peptide Profiling
- Differential Analysis of Complex Protein and Metabolite Mixtures Using Liquid Chromatography/Mass Spectrometry (LC/MS)