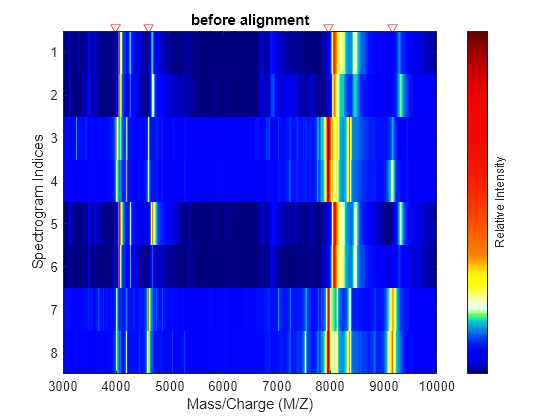
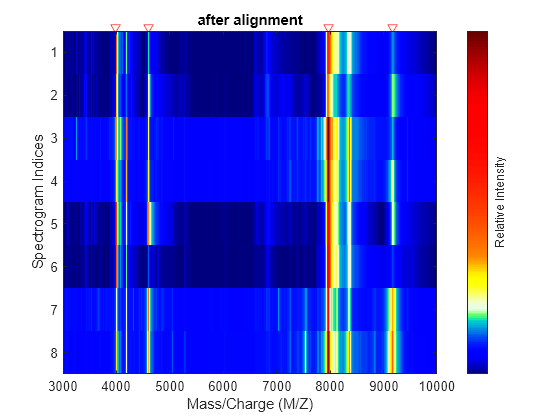
msalign
Align peaks in signal to reference peaks
Syntax
Description
IntensitiesOut = msalign(X,Intensities,RefX)Intensities and
X, to reference peaks, provided by RefX.
IntensitiesOut = msalign(X,Intensities,RefX,Name,Value)msalign using one or more
Name=Value arguments. For example, you can have msalign
take more iterations than the default five by specifying
Iterations=10.
[
also returns IntensitiesOut,RefXOut] = msalign(X,Intensities,RefX,Name,Value)RefXout, a new vector of separation-unit values to use as
reference masses for aligning the peaks. RefXOut differs from
RefX only when the Group name-value argument is
true.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Algorithms
First, msalign creates a synthetic signal from the reference peaks
using Gaussian pulses centered at the separation-unit values specified by
RefX. Then, msalign shifts and scales the
separation-unit scale to find the maximum alignment between the input signals and the synthetic
signal. (msalign uses an iterative multiresolution grid search until it
finds the best scale and shift factors for each signal.) Once msalign
determines the new separation-unit scale, msalign creates the corrected
signals by resampling their intensities at the original separation-unit values, creating
IntensitiesOut, a vector or matrix of corrected intensity values. The
resampling method preserves the shape of the peaks.
References
[1] Monchamp, P., Andrade-Cetto, L., Zhang, J.Y., and Henson, R. (2007) Signal Processing Methods for Mass Spectrometry. In Systems Bioinformatics: An Engineering Case-Based Approach, G. Alterovitz and M.F. Ramoni, eds. (Artech House Publishers).
Version History
Introduced before R2006a
See Also
mspalign | msbackadj | msdotplot | msheatmap | mslowess | msnorm | mspeaks | msresample | msppresample | mssgolay | msviewer
Topics
- Mass Spectrometry and Bioanalytics
- Preprocessing Raw Mass Spectrometry Data
- Visualizing and Preprocessing Hyphenated Mass Spectrometry Data Sets for Metabolite and Protein/Peptide Profiling
- Differential Analysis of Complex Protein and Metabolite Mixtures Using Liquid Chromatography/Mass Spectrometry (LC/MS)