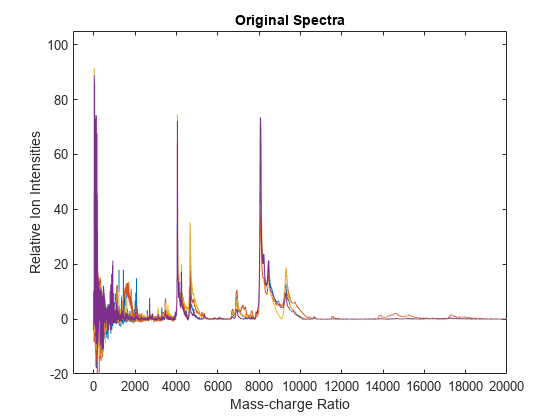
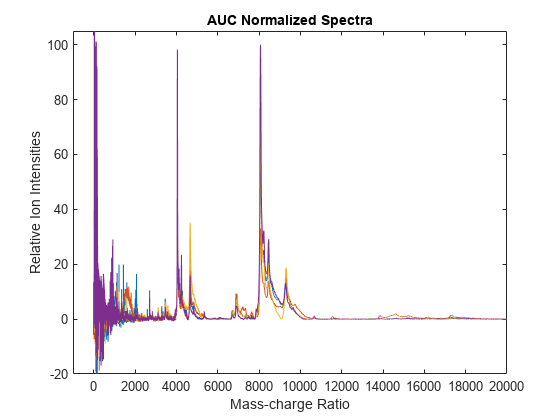
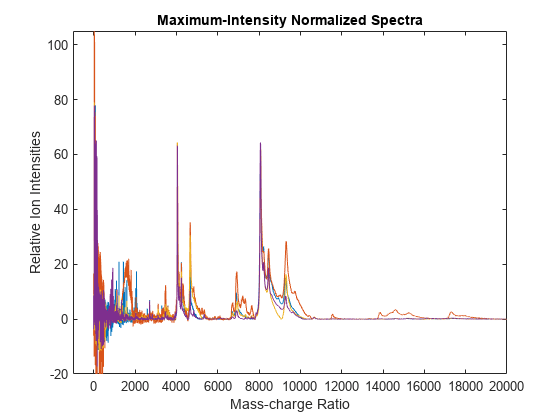
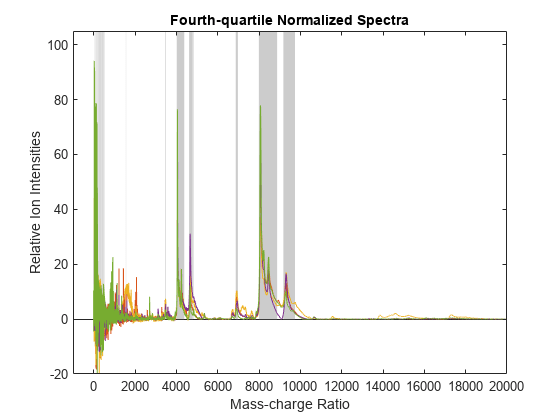
msnorm
Normalize set of signals with peaks
Syntax
Description
yOut = msnorm(X,Intensities)yOut.
[
also returns the normalization parameters yOut,normParams] = msnorm(X,Intensities)normParams, which you can use
to normalize another group of signals.
yOut = msnorm(X,Intensities,NormParameters)NormParameters from a previous
normalization to normalize a new set of signals. The function uses the same parameters to
select the separation-unit positions and output scale from the previous normalization. If
you specified a consensus proportion using the 'Consensus' name-value
pair argument in the previous normalization, the function selects no new separation-unit
positions and performs normalization using the same separation-unit positions.
[___] = msnorm(
uses additional options specified by one or more name-value pair arguments and returns any
of the output arguments in previous syntaxes. For example, X,Intensities,Name,Value)out =
msnorm(X,Y,'Quantile',[0.9 1]) sets the lower (0.9) and upper (1) quantile limit
to use only the largest 10% of intensities in each signal to compute the AUC.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Version History
Introduced before R2006a
See Also
mspalign | msbackadj | msdotplot | msalign | msheatmap | mslowess | mspeaks | msresample | msppresample | mssgolay | msviewer
Topics
- Mass Spectrometry and Bioanalytics
- Preprocessing Raw Mass Spectrometry Data
- Visualizing and Preprocessing Hyphenated Mass Spectrometry Data Sets for Metabolite and Protein/Peptide Profiling
- Differential Analysis of Complex Protein and Metabolite Mixtures Using Liquid Chromatography/Mass Spectrometry (LC/MS)