msheatmap
Create pseudocolor image of set of mass spectra
Description
msheatmap(
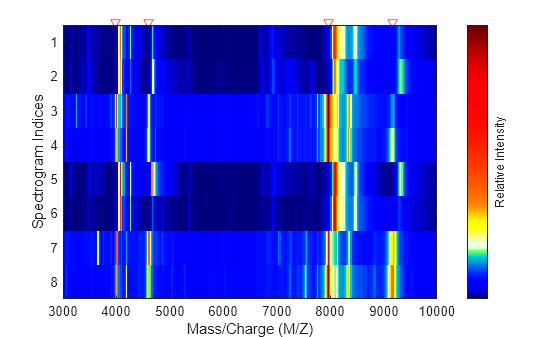
displays a pseudocolor heatmap image of the intensities for the spectra in the
MZ,Intensities)Intensities matrix.
msheatmap(
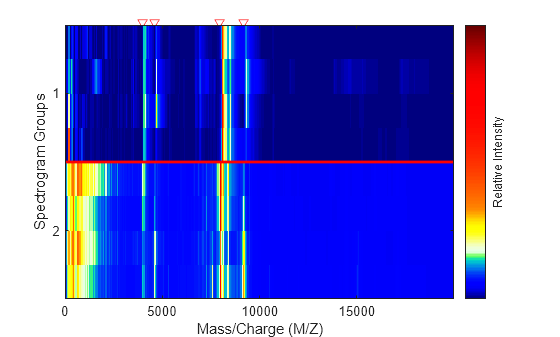
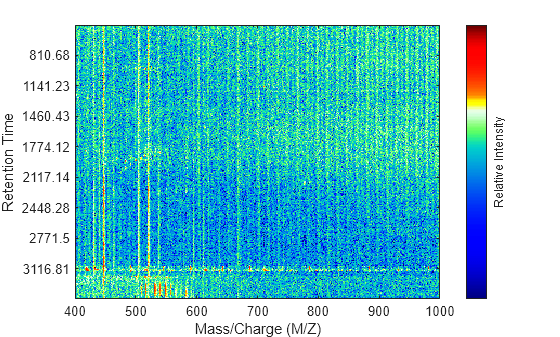
displays a pseudocolor heatmap image of the intensities for the spectra in matrix
MZ,Times,Intensities)Intensities, using the retention times in the
Times vector to label the y-axis.
msheatmap(___,
specifies additional options using one or more name-value arguments.Name=Value)
Examples
Input Arguments
Name-Value Arguments
Version History
Introduced before R2006a
See Also
mspalign | msbackadj | msdotplot | msalign | mslowess | msnorm | mspeaks | msresample | msppresample | mssgolay | msviewer
Topics
- Mass Spectrometry and Bioanalytics
- Preprocessing Raw Mass Spectrometry Data
- Visualizing and Preprocessing Hyphenated Mass Spectrometry Data Sets for Metabolite and Protein/Peptide Profiling
- Differential Analysis of Complex Protein and Metabolite Mixtures Using Liquid Chromatography/Mass Spectrometry (LC/MS)