提高浅层神经网络泛化能力,避免过拟合
提示
要了解如何为深度学习网络设置参数,请参阅trainingOptions。
神经网络训练中出现的问题之一称为过拟合。网络在训练集上的误差达到一个非常小的值、但在提交给网络的新数据上误差很大。网络记忆了训练示例,但尚未学会泛化应用于新情况。
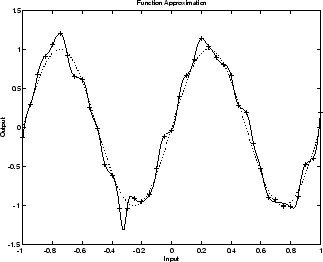
下图显示一个 1-20-1 神经网络的响应,该网络已经过训练,用于逼近含噪正弦函数。虚线表示基础正弦函数,+ 符号表示含噪测量值,实线表示神经网络响应。很明显,此网络对数据进行了过拟合,将无法很好地泛化。
提高网络泛化能力的一种方法是使用足够大的网络来提供足够的拟合度。您使用的网络越大,网络可以创建的函数就越复杂。如果您使用的网络足够小,它将没有能力对数据进行过拟合。
遗憾的是,很难预知特定应用的合适网络规模。Deep Learning Toolbox™ 软件还实现了另外两种提高泛化能力的方法:正则化和早停法。接下来的几节说明这两种方法及其实现例程。
请注意,如果网络中的参数个数远远少于训练集中的总点数,则过拟合极少发生或根本不会发生。如果您可以轻松地采集更多数据并增加训练集的大小,则不必担心以下方法会防止过拟合。本节的其余部分仅适用于您要充分利用有限数据源的情况。
重新训练神经网络
通常情况下,每个反向传播训练会话都从不同的初始权重和偏置开始,并以不同方式将数据划分为训练集、验证集和测试集。对于同一个问题,这些不同条件会导致相差很大的解。
最好训练几个网络,以确保找到具有良好泛化能力的网络。
在此处,加载数据集并将其划分为两部分:90% 用于设计网络,10% 用于测试所有网络。
[x, t] = bodyfat_dataset; Q = size(x, 2); Q1 = floor(Q * 0.90); Q2 = Q - Q1; ind = randperm(Q); ind1 = ind(1:Q1); ind2 = ind(Q1 + (1:Q2)); x1 = x(:, ind1); t1 = t(:, ind1); x2 = x(:, ind2); t2 = t(:, ind2);
接下来,选择一种网络架构并基于数据集的第一部分进行十次训练,并求出各网络架构在数据集第二部分上的均方误差。
net = feedforwardnet(10);
numNN = 10;
NN = cell(1, numNN);
perfs = zeros(1, numNN);
for i = 1:numNN
fprintf('Training %d/%d\n', i, numNN);
NN{i} = train(net, x1, t1);
y2 = NN{i}(x2);
perfs(i) = mse(net, t2, y2);
end每个网络都将从不同初始权重和偏置开始进行训练,并将第一个数据集以不同比例划分为训练集、验证集和测试集。请注意,测试集适用于每个对应网络的泛化能力,但不适合所有网络,因为作为一个网络的测试集的数据可能会被其他神经网络用于训练或验证。这也是为什么原始数据集被划分为两部分,这是为了确保保留完全独立的测试集。
性能最差的神经网络是泛化用于数据集第二部分时效果最佳的网络。
多个神经网络
提高泛化能力的另一个简单方法是训练多个神经网络并对其输出取平均值,尤其是对于含噪数据或小型数据集。
例如,此处针对一个小问题训练 10 个神经网络,并将其均方误差与平均值的均方误差进行比较。
首先,加载数据集并将其划分为设计集和测试集。
[x, t] = bodyfat_dataset; Q = size(x, 2); Q1 = floor(Q * 0.90); Q2 = Q - Q1; ind = randperm(Q); ind1 = ind(1:Q1); ind2 = ind(Q1 + (1:Q2)); x1 = x(:, ind1); t1 = t(:, ind1); x2 = x(:, ind2); t2 = t(:, ind2);
然后,训练十个神经网络。
net = feedforwardnet(10);
numNN = 10;
nets = cell(1, numNN);
for i = 1:numNN
fprintf('Training %d/%d\n', i, numNN)
nets{i} = train(net, x1, t1);
end接下来,基于第二个数据集对每个网络进行测试,得到单一网络性能和平均输出性能。
perfs = zeros(1, numNN);
y2Total = 0;
for i = 1:numNN
neti = nets{i};
y2 = neti(x2);
perfs(i) = mse(neti, t2, y2);
y2Total = y2Total + y2;
end
perfs
y2AverageOutput = y2Total / numNN;
perfAveragedOutputs = mse(nets{1}, t2, y2AverageOutput) 平均输出的均方误差可能低于大多数单一网络的性能,但未必是全部。它可能会更好地泛化应用于其他新数据。
对于一些高难度问题,可以训练 100 个网络,并对任一输入取其输出的平均值。这尤其适合小型含噪数据集与贝叶斯正则化训练函数 trainbr 结合使用的情况,如下所述。
早停法
提高泛化能力的默认方法称为早停法。该方法自动提供给所有的有监督网络创建函数,包括反向传播网络创建函数,如 feedforwardnet。
在此方法中,可用数据划分为三个子集。第一个子集是训练集,用于计算梯度和更新网络权重及偏置。第二个子集是验证集。在训练过程中会监控基于验证集的误差。验证误差通常在训练的初始阶段减小,训练集误差也是如此。然而,当网络开始过拟合数据时,基于验证集的误差通常开始增大。当验证误差在指定次数的迭代 (net.trainParam.max_fail) 中增加时,训练停止,并返回验证误差最小时的权重和偏置。
训练期间不使用测试集误差,但测试集误差用于比较不同模型。在训练过程中绘制测试集误差也很有用。如果测试集误差与验证集误差达到最小值所需的迭代次数显著不同,这可能表示数据集的划分不佳。
可使用四个函数将数据划分为训练集、验证集和测试集。它们是 dividerand(默认值)、divideblock、divideint 和 divideind。您可以使用以下属性访问或更改网络的划分函数:
net.divideFcn
其中每个函数都采用自定义其行为的参数。这些值会被存储,且可以通过以下网络属性进行更改:
net.divideParam
索引数据划分 (divideind)
创建一个简单的测试问题。对于完整的数据集,以 0.01 的步长生成有 201 个输入点的含噪正弦波,输入点范围为 −1 至 1:
p = [-1:0.01:1]; t = sin(2*pi*p)+0.1*randn(size(p));
按索引划分数据,以便将连续样本依次分配给训练集、验证集和测试集:
trainInd = 1:3:201 valInd = 2:3:201; testInd = 3:3:201; [trainP,valP,testP] = divideind(p,trainInd,valInd,testInd); [trainT,valT,testT] = divideind(t,trainInd,valInd,testInd);
随机数据划分 (dividerand)
您可以随机划分输入数据,以便将 60% 的样本分配给训练集,20% 分配给验证集,20% 分配给测试集,如下所示:
[trainP,valP,testP,trainInd,valInd,testInd] = dividerand(p);
该函数不仅划分输入数据,还返回索引,以便您可以使用 divideind 相应地划分目标数据:
[trainT,valT,testT] = divideind(t,trainInd,valInd,testInd);
分块数据划分 (divideblock)
您还可以随机划分输入数据,以便将前 60% 的样本分配给训练集,接下来的 20% 分配给验证集,最后 20% 分配给测试集,如下所示:
[trainP,valP,testP,trainInd,valInd,testInd] = divideblock(p);
使用 divideind 相应地划分目标数据:
[trainT,valT,testT] = divideind(t,trainInd,valInd,testInd);
交错数据划分 (divideint)
划分输入数据的另一种方法是根据百分比将样本循环分配给训练集、验证集和测试集。您可以按照交错方式将 60% 的样本分配给训练集、20% 分配给验证集、20% 分配给测试集,如下所示:
[trainP,valP,testP,trainInd,valInd,testInd] = divideint(p);
使用 divideind 相应地划分目标数据。
[trainT,valT,testT] = divideind(t,trainInd,valInd,testInd);
正则化
另一种提高泛化能力的方法称为正则化。这涉及到修正性能函数,通常选择性能函数来作为基于训练集的网络误差的平方和。下一节解释如何修正性能函数,其后的一节说明自动设置最优性能函数以实现最佳泛化的例程。
修正的性能函数
用于训练前馈神经网络的典型性能函数是网络误差的均值平方和。
如果您通过添加由网络权重和偏置的平方和均值组成的项 来修正性能函数,则可能会提高泛化能力;其中 是性能比,并且
使用此性能函数会减小网络的权重和偏置,这将迫使网络响应更平滑,降低过拟合的可能性。
以下代码重新初始化以前的网络,并使用 BFGS 算法和正则化性能函数对其进行重新训练。此处,性能比设置为 0.5,这使得均方误差和均方权重具有相等的权重。
[x,t] = simplefit_dataset; net = feedforwardnet(10,'trainbfg'); net.divideFcn = ''; net.trainParam.epochs = 300; net.trainParam.goal = 1e-5; net.performParam.regularization = 0.5; net = train(net,x,t);
正则化的问题是很难确定性能比参数的最优值。如果将此参数设置得太大,可能会出现过拟合。如果该比率太小,则网络无法充分拟合训练数据。下一节说明自动设置正则化参数的例程。
自动正则化 (trainbr)
该方法以自动方式确定最佳的正则化参数。此过程使用的一种方法是 David MacKay 提出的贝叶斯框架 [MacK92]。在此框架中,网络的权重和偏置假设为服从指定分布的随机变量。正则化参数与这些分布的未知方差相关。然后,您可以使用统计方法来估计这些参数。
贝叶斯正则化的详细讨论超出了本用户指南的范围。有关结合使用贝叶斯正则化和莱文贝格-马夸特训练的详细说明,请参阅 [FoHa97]。
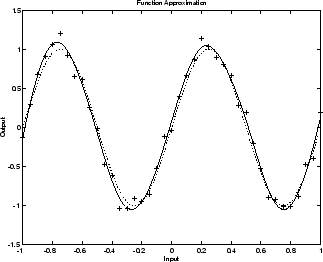
贝叶斯正则化已通过函数 trainbr 实现。以下代码说明如何使用此函数来训练 1-20-1 网络,以逼近提高浅层神经网络泛化能力,避免过拟合的图中所示的含噪正弦波。(通过设置 net.divideFcn 取消数据划分,从而独立于早停法分析 trainbr 的效果。)
x = -1:0.05:1; t = sin(2*pi*x) + 0.1*randn(size(x)); net = feedforwardnet(20,'trainbr'); net = train(net,x,t);
该算法的一个特点是,它可以测量网络实际上在使用多少个网络参数(权重和偏置)。在本例中,最终经过训练的网络使用 1-20-1 网络中的 61 个总权重和偏置中的大约 12 个参数(由打印输出中的 #Par 指示)。不管网络中的参数个数变得多大,此实际参数个数应保持大致相同。(此论述假设网络已训练足够的迭代次数以确保收敛。)
trainbr 算法通常在网络输入和目标缩放到大致在 [−1,1] 范围内时效果最好。此处的测试问题就是这种情况。如果您的输入和目标不在此范围内,可以使用函数 mapminmax 或 mapstd 来执行缩放,如选择神经网络输入输出处理函数中所述。默认情况下,使用 feedforwardnet 创建的网络包含 mapminmax 作为输入和输出处理函数。
下图显示经过训练的网络的响应。与上一个图(图中 1-20-1 网络过拟合数据)相比,您可以看到网络响应非常接近基础正弦函数(虚线),因此,网络对新输入具有很强的泛化能力。您可以尝试更大的网络,但网络响应永远不会过拟合数据。这就消除了确定最优网络大小所需的猜测工作。
当使用 trainbr 时,务必让算法运行到实际参数个数收敛为止。训练可能会停止并显示“Maximum MU reached”消息,这是典型情形,明确表示算法已真正收敛。如果误差平方和 (SSE) 和权重平方和 (SSW) 在几次迭代中相对恒定,则也可以视为算法已收敛。出现这种情况时,您可能需要点击训练窗口中的停止按钮。
早停法和正则化的摘要与讨论
如果应用得当,早停法和正则化可以确保网络泛化。
对于早停法,您必须小心,不要使用收敛太快的算法。如果您使用快速算法(如 trainlm),请合理设置训练参数,使收敛相对较慢。例如,将 mu 设置为相对较大的值(如 1),并将 mu_dec 和 mu_inc 分别设置为接近 1 的值(如 0.8 和 1.5)。训练函数 trainscg 和 trainbr 通常在早停法中运行良好。
使用早停法时,验证集的选择也很重要。验证集应能代表训练集中的所有点。
当使用贝叶斯正则化时,务必将网络训练到收敛为止。当网络收敛时,误差平方和、权重平方和以及有效参数个数应达到恒定值。
使用早停法和正则化时,最好从几个不同初始条件开始训练网络。在某些情况下,其中一种方法可能失败。通过测试几种不同初始条件,您可以确认稳健网络性能。
当数据集很小并且您正在训练函数逼近网络时,贝叶斯正则化可提供比早停法更好的泛化性能。这是因为贝叶斯正则化不要求验证数据集与训练数据集分离;它使用所有数据。
为了深入了解算法的性能,我们在几个基准数据集上测试了早停法和贝叶斯正则化,如下表所示。
数据集标题 | 点数 | 网络 | 描述 |
|---|---|---|---|
BALL | 67 | 2-10-1 | 小球位置测量中的双传感器标定 |
SINE (5% N) | 41 | 1-15-1 | 高斯噪声水平为 5% 的单周期正弦波 |
SINE (2% N) | 41 | 1-15-1 | 高斯噪声水平为 2% 的单周期正弦波 |
ENGINE (ALL) | 1199 | 2-30-2 | 发动机传感器 - 完整数据集 |
ENGINE (1/4) | 300 | 2-30-2 | 发动机传感器 - 数据集的 1/4 |
CHOLEST (ALL) | 264 | 5-15-3 | 胆固醇测量 - 完整数据集 |
CHOLEST (1/2) | 132 | 5-15-3 | 胆固醇测量 - 1/2 数据集 |
这些数据集有不同的大小以及不同数量的输入和目标。使用其中两个数据集,使用所有数据对网络进行一次训练,然后仅使用一部分数据重新训练。这说明当数据集较小时,贝叶斯正则化的优势变得更加明显。除了 SINE 数据集以外,所有数据集都是从物理系统获得的。这两个数据集是通过在正弦波的单个周期中添加不同水平的噪声而有意创建的。算法在这两个数据集上的性能说明了噪声的影响。
下表摘要显示了在七个测试集上早停法和贝叶斯正则化的性能。(trainscg 算法用于早停法测试。其他算法提供类似的性能。)
均方测试集误差
| 方法 | Ball | Engine (All) | Engine (1/4) | Choles (All) | Choles (1/2) | Sine (5% N) | Sine (2% N) |
|---|---|---|---|---|---|---|---|
| ES | 1.2e-1 | 1.3e-2 | 1.9e-2 | 1.2e-1 | 1.4e-1 | 1.7e-1 | 1.3e-1 |
| BR | 1.3e-3 | 2.6e-3 | 4.7e-3 | 1.2e-1 | 9.3e-2 | 3.0e-2 | 6.3e-3 |
| ES/BR | 92 | 5 | 4 | 1 | 1.5 | 5.7 | 21 |
您可以看到,在大多数情况下,贝叶斯正则化的性能优于早停法。当数据集很小时,或当数据集中几乎没有噪声时,性能的改善最为明显。例如 BALL 数据集,它是从噪声很小的传感器中获得的。
尽管贝叶斯正则化的泛化性能通常优于早停法,但并非始终如此。此外,在工具箱中实现的贝叶斯正则化形式在模式识别问题上的性能不如在函数逼近问题上。这是因为当网络输出饱和时,在莱文贝格-马夸特算法中使用的对黑塞矩阵的逼近不如在模式识别问题中准确。贝叶斯正则化方法的另一个缺点是,它通常比早停法花费更长的时间才能收敛。
训练后分析 (regression)
经过训练的网络的性能在一定程度上可以通过基于训练集、验证集和测试集的误差来测量,但更详细地调查网络响应通常是有用的。一种选择是执行网络响应和对应目标之间的回归分析。例程 regression 就是为执行此分析而设计的。
以下命令说明如何对经过训练的网络执行回归分析。
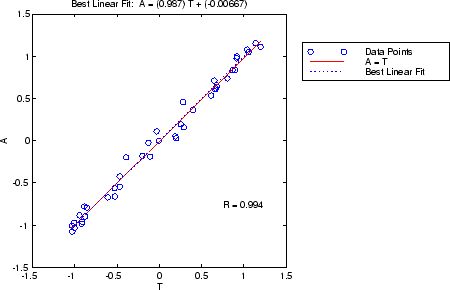
x = [-1:.05:1]; t = sin(2*pi*x)+0.1*randn(size(x)); net = feedforwardnet(10); net = train(net,x,t); y = net(x); [r,m,b] = regression(t,y)
r =
0.9935
m =
0.9874
b =
-0.0067
将网络输出和对应的目标传递给 regression。它返回三个参数。前两个参数 m 和 b 对应于使目标与网络输出具有最佳线性回归的斜率和 y 截距。如果存在完美拟合(输出完全等于目标),则斜率为 1,y 截距为 0。在此示例中,您可以看到数值非常接近它们。regression 返回的第三个变量是输出和目标之间的相关系数(R 值)。它用于衡量目标解释输出变化的程度。如果此数值等于 1,则目标和输出之间就有完美的相关性。在此示例中,该数值非常接近 1,这表明拟合良好。
下图显示 regression 提供的图形输出。它是网络输出(用空心圆表示)对目标的图。最佳线性拟合由虚线表示。实线表示完美拟合(输出等于目标)。在此示例中,由于拟合非常好,很难区分最佳线性拟合线和完美拟合线。