使用深度学习进行“序列到单个”回归
此示例说明如何使用长短期记忆 (LSTM) 神经网络来预测波形的频率。
您可以使用 LSTM 神经网络,通过序列和目标值的训练集来预测序列的数值响应。LSTM 网络是一种循环神经网络 (RNN),它通过遍历时间步并更新网络状态来处理输入数据。网络状态包含在先前时间步中记住的信息。序列的数值响应示例包括:
序列的属性,如频率、最大值和均值。
序列的过去或将来时间步的值。
此示例使用 Waveform 数据集训练“序列到单个”回归 LSTM 网络,该数据集包含生成的 1000 个不同长度的合成波形,有三个通道。要使用传统方法确定波形的频率,请参阅fft。
加载序列数据
从 WaveformData.mat 加载示例数据。数据是序列的 numObservations×1 元胞数组,其中 numObservations 是序列数。每个序列都是一个 numTimeSteps×numChannels 数值数组,其中 numTimeSteps 是序列中的时间步数,numChannels 是序列的通道数。对应的目标位于波形频率的 numObservations×numResponses 数值数组中,其中 numResponses 是目标的通道数。
load WaveformData查看观测值数目。
numObservations = numel(data)
numObservations = 1000
查看前几个序列的大小和对应的频率。
data(1:4)
ans=4×1 cell array
{103×3 double}
{136×3 double}
{140×3 double}
{124×3 double}
freq(1:4,:)
ans = 4×1
5.8922
2.2557
4.5250
4.4418
查看序列的通道数。对于网络训练,每个序列必须具有相同数量的通道。
numChannels = size(data{1},2)numChannels = 3
查看响应的数量(目标的通道数量)。
numResponses = size(freq,2)
numResponses = 1
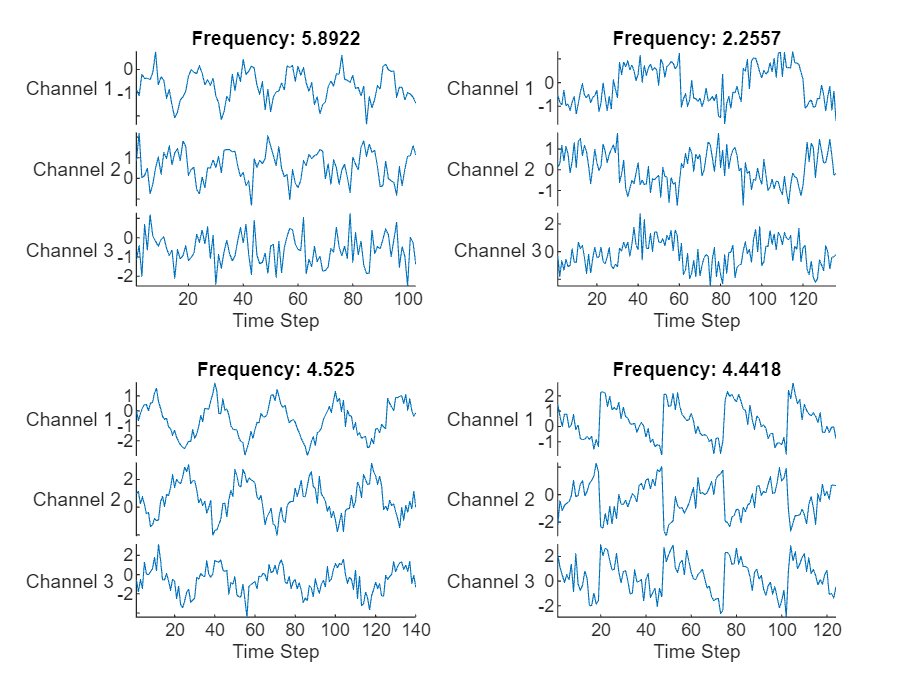
可视化绘图中的前几个序列。
figure tiledlayout(2,2) for i = 1:4 nexttile stackedplot(data{i}, DisplayLabels="Channel " + (1:numChannels)) xlabel("Time Step") title("Frequency: " + freq(i)) end
准备要训练的数据
留出用于验证和测试的数据。将数据划分为训练集(包含 80% 的数据)、验证集(包含 10% 的数据)和测试集(包含其余 10% 的数据)。
[idxTrain,idxValidation,idxTest] = trainingPartitions(numObservations, [0.8 0.1 0.1]); XTrain = data(idxTrain); XValidation = data(idxValidation); XTest = data(idxTest); TTrain = freq(idxTrain); TValidation = freq(idxValidation); TTest = freq(idxTest);
定义 LSTM 网络架构
创建 LSTM 回归网络。
使用输入大小与输入数据的通道数匹配的序列输入层。
为了更好地拟合并防止训练发散,请将序列输入层的
Normalization选项设置为“zscore”。这会将序列数据归一化为具有零均值和单位方差。接下来,使用一个具有 100 个隐藏单元的 LSTM 层。隐藏单元的数量确定该层学习了多少信息。较大的值可以产生更准确的结果,但也容易导致训练数据过拟合。
要为每个序列输出一个时间步,请将 LSTM 层的
OutputMode选项设置为“last”。要指定要预测的值的数目,请包括一个大小与预测变量值数目匹配的全连接层。
numHiddenUnits = 100; layers = [ ... sequenceInputLayer(numChannels, Normalization="zscore") lstmLayer(numHiddenUnits, OutputMode="last") fullyConnectedLayer(numResponses)]
layers =
3×1 Layer array with layers:
1 '' Sequence Input Sequence input with 3 dimensions
2 '' LSTM LSTM with 100 hidden units
3 '' Fully Connected 1 fully connected layer
指定训练选项
指定训练选项。在选项中进行选择需要经验分析。要通过运行试验探索不同训练选项配置,您可以使用Experiment Manager。
使用 Adam 优化器进行训练。
进行 250 轮训练。对于较大的数据集,您可能不需要像良好拟合那样进行这么多轮训练。
指定用于验证的序列和响应。
输出给出最佳(即最低)验证损失的网络。
将学习率设置为 0.005。
截断每个小批量中的序列,使其长度与最短的序列相同。截断序列可确保不添加任何填充,但代价是丢弃数据。对于序列中的所有时间步都可能包含重要信息的序列,截断将使网络无法实现良好的拟合。
在图中监控训练进度并监控 RMSE 度量。
禁用详尽输出。
options = trainingOptions("adam", ... MaxEpochs=250, ... ValidationData={XValidation TValidation}, ... InitialLearnRate=0.005, ... SequenceLength="shortest", ... Metrics="rmse", ... Plots="training-progress", ... Verbose=false);
训练 LSTM 网络
使用 trainnet 函数训练神经网络。对于回归,请使用均方误差损失。默认情况下,trainnet 函数使用 GPU(如果有)。使用 GPU 需要 Parallel Computing Toolbox™ 许可证和受支持的 GPU 设备。有关受支持设备的信息,请参阅GPU 计算要求 (Parallel Computing Toolbox)。否则,该函数使用 CPU。要指定执行环境,请使用 ExecutionEnvironment 训练选项。
net = trainnet(XTrain,TTrain,layers,"mse",options);
测试 LSTM 网络
使用 minibatchpredict 函数进行预测。默认情况下,minibatchpredict 函数使用 GPU(如果有)。
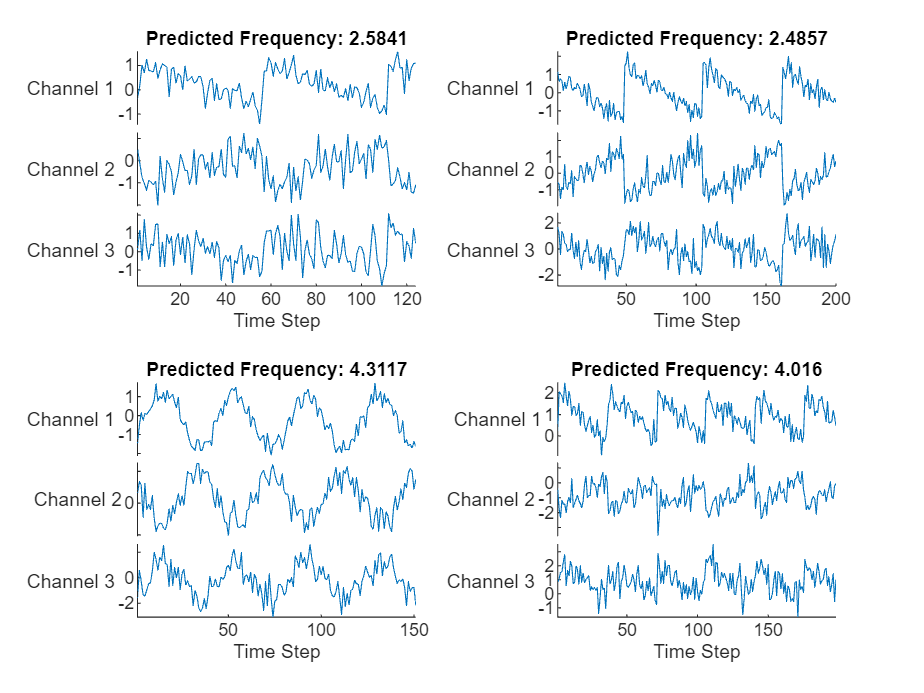
YTest = minibatchpredict(net,XTest,SequenceLength="shortest");可视化绘图中的前几个预测。
figure tiledlayout(2,2) for i = 1:4 nexttile stackedplot(XTest{i},DisplayLabels="Channel " + (1:numChannels)) xlabel("Time Step") title("Predicted Frequency: " + string(YTest(i))) end
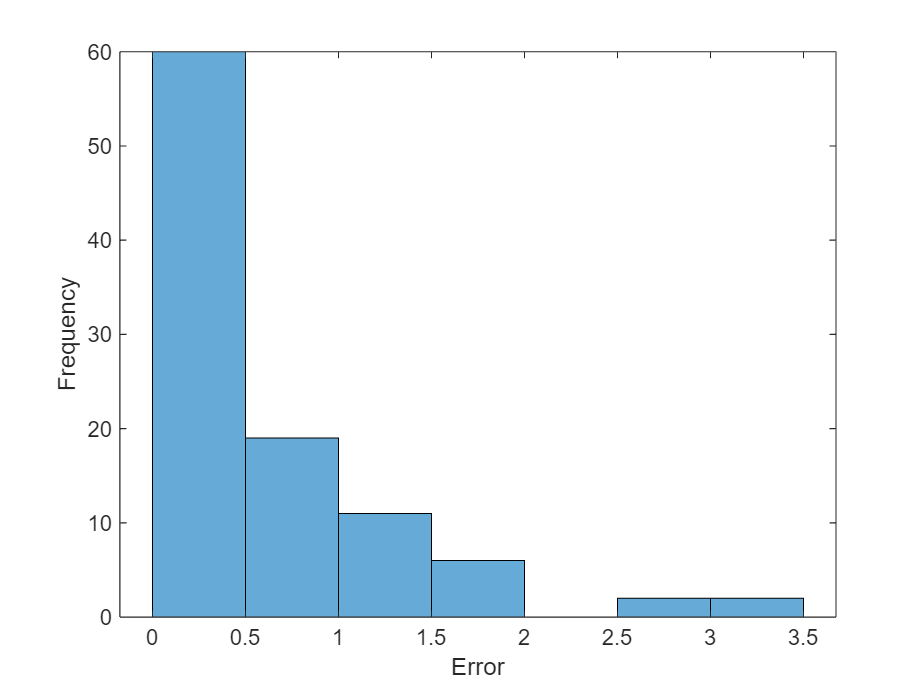
在直方图中可视化均方误差。
figure histogram(mean((TTest - YTest).^2,2)) xlabel("Error") ylabel("Frequency")
计算总体均方根误差。
rmse = rmse(YTest,TTest)
rmse = single
0.7605
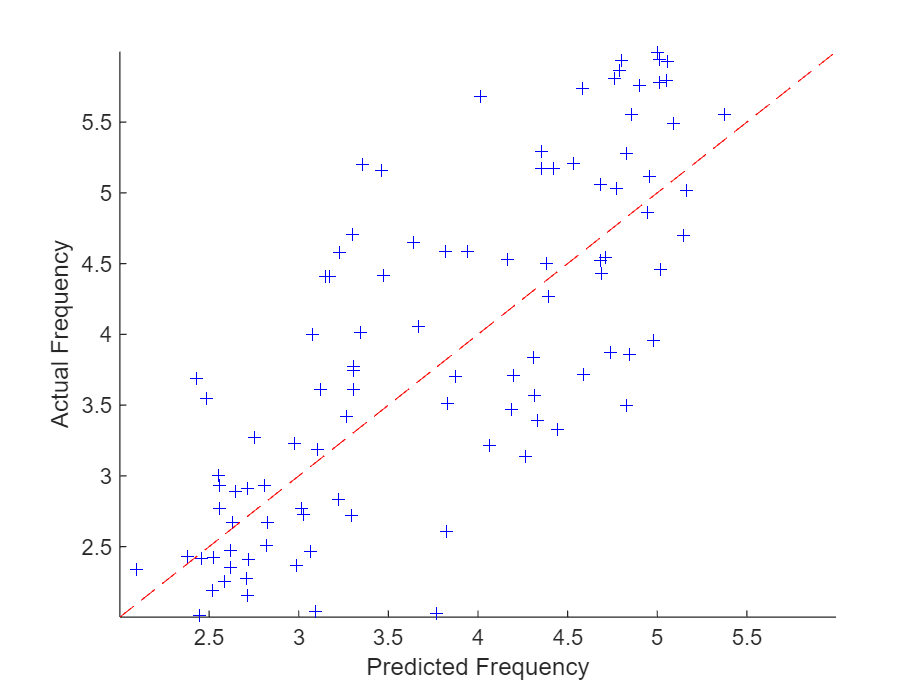
绘制预测频率对实际频率的图。
figure scatter(YTest,TTest, "b+"); xlabel("Predicted Frequency") ylabel("Actual Frequency") hold on m = min(freq); M=max(freq); xlim([m M]) ylim([m M]) plot([m M], [m M], "r--")
另请参阅
trainnet | trainingOptions | dlnetwork | testnet | minibatchpredict | scores2label | predict | lstmLayer | sequenceInputLayer