Aerial Lidar Semantic Segmentation Using RandLANet Deep Learning
This example shows how to train a RandLANet deep learning network to perform semantic segmentation on aerial lidar data.
You can use lidar data acquired from airborne laser scanning systems in applications such as topographic mapping, city modeling, biomass measurement, and disaster management. Extracting meaningful information from this data requires semantic segmentation, a process in which you assign each point in the point cloud a unique class label.
In this example, you train a RandLANet [1] network to perform semantic segmentation by using the Dayton Annotated Lidar Earth Scan (DALES) data set [2]. The data set contains scenes of dense, labeled aerial lidar data from urban, suburban, rural, and commercial settings. The data set provides semantic segmentation labels for eight classes: buildings, cars, trucks, poles, power lines, fences, ground, and vegetation.
Load DALES Data
The DALES data set contains 40 scenes of aerial lidar data. Out of the 40 scenes, 29 scenes are used for training and the remaining 11 scenes are used for testing. Follow the instructions on the DALES website to download the data set to the folder specified by the dataFolder variable. Create folders to store the training and test data.
dataFolder = fullfile(tempdir,"DALES"); trainDataFolder = fullfile(dataFolder,"dales_las","train"); testDataFolder = fullfile(dataFolder,"dales_las","test");
Preview a point cloud from the training data.
lasReader = lasFileReader(fullfile(trainDataFolder,"5080_54435.las")); [pc,attr] = readPointCloud(lasReader,"Attributes","Classification"); labels = attr.Classification; % Select only labeled data. pc = select(pc,labels~=0); labels = labels(labels~=0); classNames = ["ground"; ... "vegetation"; ... "cars"; ... "trucks"; ... "powerlines"; ... "fences"; ... "poles"; ... "buildings"]; figure ax = pcshow(pc.Location,labels); helperLabelColorbar(ax,classNames) title("Point Cloud with Overlaid Semantic Labels")

Preprocess Training Data
To preprocess the training data, first create a fileDatastore object to collect all the point cloud files in the training data.
fds = fileDatastore(trainDataFolder,"ReadFcn",@lasFileReader);Each point cloud in the DALES data set covers an area of 500-by-500 meters containing around 10 millions points, processing this many points might slow down network processing. For efficient memory processing, divide the point cloud into small, non-overlapping blocks of size 50-by-50 meters by using the helperTransformData helper function, defined at the end of this example.
Define the block dimensions.
blockSize = [50 50];
Apply the transformation to the training datastore.
ldsTransformed = transform(fds,@(x) helperTransformData(x,blockSize));
Use the helperDatastoreWriteFcn helper function, attached to this example as a supporting file, to save the preprocessed point clouds and semantic labels as PCD and PNG files, respectively. You can set writeFiles to false if your preprocessed training data is already present in the outputFolder.
writeFiles = true; outputFolder = fullfile(dataFolder,"PreprocessedTrainData"); if writeFiles writeall(ldsTransformed,outputFolder,WriteFcn = @helperDatastoreWriteFcn,FolderLayout = "flatten"); end
Processing done for file: 5190_54400.las
Note: Processing can take some time. The code suspends MATLAB® execution until processing is complete.
If you want to rerun the code, you must first use the "clear helperDatastoreWriteFcn" command.
Create Datastore Object for Training
Create a fileDatastore object that reads PCD files by using the pcread function.
pcCropTrainPath = fullfile(outputFolder,"PointCloud"); ldsTrain = fileDatastore(pcCropTrainPath,"ReadFcn",@(x) pcread(x));
Use a pixelLabelDatastore object to store pixel-wise labels from the pixel label images. The object maps each pixel label to a class name and assigns a unique label ID to each class.
% Specify label IDs from 1 to the number of classes. numClasses = numel(classNames); labelIDs = 1 : numClasses; labelCropTrainPath = fullfile(outputFolder,"Labels"); pxdsTrain = pixelLabelDatastore(labelCropTrainPath,classNames,labelIDs);
Use the combine function to combine the point clouds and labels into a single datastore for training.
cds = combine(ldsTrain,pxdsTrain);
Create RandLANet Object
RandLANet is a popular neural network used for semantic segmentation of lidar point clouds. This network uses an encoder-decoder architecture with skip connections. You feed the input to a shared multilayer perceptron layer, followed by four encoding and decoding layers, to learn features for each point. At end, three fully connected layers and a dropout layer associate each point in a 3-D point cloud with a class label, such as car, truck, ground, or vegetation.
This figure shows the architecture of a RandLANet, where:
FC — Fully Connected Layer
LFA — Local Feature Aggregation
RS — Random Sampling
MLP — Shared Multilayer Perceptron
US — Upsampling
DP — Dropout

Each encoding layer randomly downsamples the point cloud, which significantly decreases the point density. To retain the key features of the downsampled point cloud, the network uses a local feature aggregation module on each point.
Each decoding layer upsamples the point feature set through nearest-neighbor interpolation. The network concatenates these upsampled maps with feature maps produced by the encoding layers through skip connections, and applies a shared MLP to the concatenated feature maps.
Use the randlanet function to create a RandLANet segmentation network.
segmenter = randlanet("none",classNames);Specify Training Options
Use the Adam optimization algorithm to train the network. Use the trainingOptions (Deep Learning Toolbox) function to specify the hyperparameters.
learningRate = 0.01; numEpochs = 200; miniBatchSize = 8; learnRateDropFactor = 0.9886; executionEnvironment = "auto"; if canUseParallelPool dispatchInBackground = true; else dispatchInBackground = false; end options = trainingOptions("adam", ... InitialLearnRate = learningRate, ... MaxEpochs = numEpochs, ... MiniBatchSize = miniBatchSize, ... LearnRateSchedule = "piecewise", ... LearnRateDropPeriod = 1,... LearnRateDropFactor = learnRateDropFactor, ... Plots = "training-progress", ... ExecutionEnvironment = executionEnvironment, ... dispatchInBackground = dispatchInBackground, ... ResetInputNormalization = false, ... CheckpointFrequencyUnit = "epoch", ... CheckpointFrequency = 10, ... CheckpointPath = tempdir, ... BatchNormalizationStatistics="moving");
Note: Reduce or increase the miniBatchSize value to control memory usage when training.
Train Model
Use the trainRandlanet function to train the RandLANet segmentation network, set the doTraining argument to true. Otherwise, load a pretrained segmentation network. To train the network, you can use a CPU or GPU. Using a GPU requires Parallel Computing Toolbox™ and a CUDA® enabled NVIDIA® GPU. For more information, see GPU Computing Requirements (Parallel Computing Toolbox).
doTraining = false; if doTraining segmenter = trainRandlanet(cds,segmenter,options); else segmenter = randlanet("dales"); end
Segment Aerial Point Cloud
Create a fileDatastore object to read multiple test point clouds. For demonstration purpose, we are reading a single point cloud.
fdsTest = fileDatastore(fullfile(testDataFolder,"5080_54470.las"),"ReadFcn",@lasFileReader);
Apply the similar transformation used on training data, to the test data:
Extract the point cloud and the respective labels.
Crop the point clouds into non-overlapping blocks of size 50-by-50 meters.
ldsTestTransformed = transform(fdsTest,@(x) helperTransformData(x,blockSize));
Read all blocks of the test point cloud. Notice X contain pointCloud objects and corresponding labels for all the blocks of the test point cloud.
X = read(ldsTestTransformed);
Create pointCloud object containing array of block of pointClouds of size Mx1. Notice segmentObjects only take pointCloud array of size Mx1 as input.
ptCloudArray = vertcat(X{:,1});Using segmentObjects function, perform inference to compute prediction labels.
[labels,scores] = segmentObjects(segmenter,ptCloudArray);
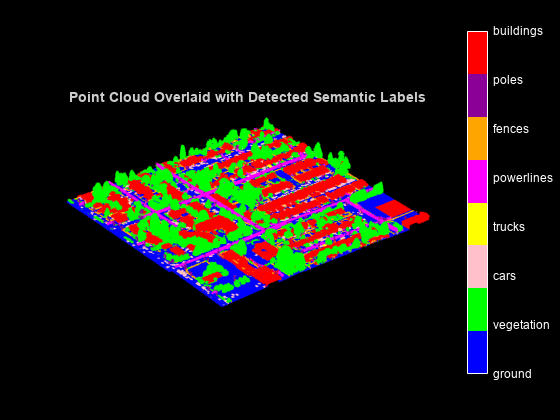
Display the predictions on the point cloud.
% Concatenate blocks of point cloud. pc = pccat(ptCloudArray); % Concatenate corresponding labels of blocks of point cloud. labelsDensePred = vertcat(labels{:}); % Convert labels from categorical to numeric for display. labelsDensePred = single(categorical(labelsDensePred,classNames,cellstr(string(1:numClasses)))); figure ax = pcshow(pc.Location,labelsDensePred); axis off helperLabelColorbar(ax,classNames) title("Point Cloud Overlaid with Detected Semantic Labels")
Evaluate Network
To perform evaluation on the test data, get the labels from the test point cloud. Use the predicted labels you computed and are stored in labelsDensePred.
Extract the ground truth labels.
labelsDenseTarget = vertcat(X{:,2});Use the evaluateSemanticSegmentation function to compute the semantic segmentation metrics from the test set results.
confusionMatrix = segmentationConfusionMatrix(double(labelsDensePred), ...
double(labelsDenseTarget),Classes = 1:numClasses);
metrics = evaluateSemanticSegmentation({confusionMatrix},classNames,Verbose = false);You can measure the amount of overlap per class using the intersection-over-union (IoU) metric.
The evaluateSemanticSegmentation function returns metrics for the entire data set, for individual classes, and for each test image. To see the metrics at the data set level, inspect the metrics.DataSetMetrics property.
metrics.DataSetMetrics
ans=1×4 table
GlobalAccuracy MeanAccuracy MeanIoU WeightedIoU
______________ ____________ _______ ___________
0.95846 0.8537 0.70479 0.92468
The data set metrics provide a high-level overview of network performance. To see the impact each class has on the overall performance, view the metrics for each class by inspecting the metrics.ClassMetrics property.
metrics.ClassMetrics
ans=8×2 table
Accuracy IoU
________ _______
ground 0.97714 0.95606
vegetation 0.91161 0.89449
cars 0.95408 0.73238
trucks 0.34457 0.23184
powerlines 0.90194 0.85462
fences 0.89157 0.48109
poles 0.87155 0.56866
buildings 0.97715 0.91921
Supporting Functions
The helperTransformData function applies these transformations to the data:
Extract the point cloud and the respective labels.
Each point cloud in the DALES data set covers an area of 500-by-500 meters, which is much larger than the typical area covered by terrestrial rotating lidar point clouds. For efficient memory processing, divide the point cloud into small, non-overlapping blocks of size 50-by-50 meters.
function out = helperTransformData(lasReader,blocksize) [ptCloud,attr] = readPointCloud(lasReader,"Attributes","Classification"); labels = attr.Classification; % Select only labeled data. ptCloud = select(ptCloud,labels~=0); labels = labels(labels~=0); % Block the input point cloud. numGridsX = round((diff(ptCloud.XLimits)+eps)/blocksize(1)); numGridsY = round((diff(ptCloud.YLimits)+eps)/blocksize(2)); [~,~,~,indx,indy] = histcounts2(ptCloud.Location(:,1),ptCloud.Location(:,2), ... [numGridsX, numGridsY],XBinLimits = ptCloud.XLimits, YBinLimits = ptCloud.YLimits); ind = sub2ind([numGridsX, numGridsY],indx,indy); out = cell(numGridsX*numGridsY,2); for num = 1:numGridsX*numGridsY idx = ind == num; if(any(idx)) out{num,1} = select(ptCloud,idx); out{num,2} = labels(idx); end end end
References
[1] Hu, Qingyong, Bo Yang, Linhai Xie, Stefano Rosa, Yulan Guo, Zhihua Wang, Niki Trigoni, and Andrew Markham. “RandLA-Net: Efficient Semantic Segmentation of Large-Scale Point Clouds.” In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 11105–14. Seattle, WA, USA: IEEE, 2020. https://doi.org/10.1109/CVPR42600.2020.01112.
[2] Varney, Nina, Vijayan K. Asari, and Quinn Graehling. “DALES: A Large-Scale Aerial LiDAR Data Set for Semantic Segmentation.” In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 717–26. Seattle, WA, USA: IEEE, 2020. https://doi.org/10.1109/CVPRW50498.2020.00101.
See Also
Objects
Functions
Apps
- Deep Network Designer (Deep Learning Toolbox) | Lidar Viewer | Lidar Labeler