Deformable Registration Using Medical Registration Estimator App
This example uses a deformable registration technique supported by the Medical Registration Estimator app to estimate the displacement field between two CT scans acquired during inhalation and exhalation for the same patient.
Deformable image registration aligns 3-D volumes while accounting for non-rigid motion, such as expanding and contracting lung tissue. Deformable registration of CT scans acquired during inhalation and exhalation can map the displacement of lung tissue during the two phases of breathing. You can use the Medical Registration Estimator app to perform 3-D image registration interactively, with visual feedback. To learn more about the app, see Get Started with Medical Registration Estimator.
For a version of this example that performs registration outside the app using the imregdeform function, see Estimate Lung Displacement Field During Breathing Using Deformable Image Registration.
Download Data
This example uses a subset of data from one subject in a data set containing CT and PET images [1][2]. The subset includes one CT scan acquired during inhalation and one CT scan acquired during exhalation. The size of the subset of data is approximately 137 MB. Run this code to download the data set from the MathWorks® website, and then unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical", ... "CT-PET-Ventilation-Imaging/Chest-Ventilation-Imaging.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
After you run the code to download the data, the dataFolder folder contains the downloaded and unzipped data. Each scan consists of a directory of DICOM files.
dataFolder = fullfile(filepath,"Chest-Ventilation-Imaging");Import Data
Read the CT volume acquired at 80% of maximum inhalation as a medicalVolume object. The voxel spacing is stored in the VoxelSpacing property.
medVolInsp = medicalVolume(fullfile(dataFolder,"5.000000-Thorax Insp 2.0 B70f-84301"))medVolInsp =
medicalVolume with properties:
Voxels: [512×512×167 int16]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [0.9766 0.9766 1.8000]
NormalVector: [0 0 1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 167
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "CT"
WindowCenters: []
WindowWidths: []
Read the CT volume acquired at peak exhalation as a medicalVolume object.
medVolExsp = medicalVolume(fullfile(dataFolder,"7.000000-Thorax Exp 2.0 B70f-98860"))medVolExsp =
medicalVolume with properties:
Voxels: [512×512×167 int16]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [0.9766 0.9766 1.8000]
NormalVector: [0 0 1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 167
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "CT"
WindowCenters: []
WindowWidths: []
Open Medical Registration Estimator
Open the Medical Registration Estimator app by using the medicalRegistrationEstimator function, specifying the volumes as inputs. You can also open the app from the Apps tab on the MATLAB® toolstrip and manually import the volumes. See Get Started with Medical Registration Estimator for details.
medicalRegistrationEstimator(medVolInsp,medVolExsp)
The app opens and displays the volumes as 2-D slice overlays in each anatomical plane and in 3-D.
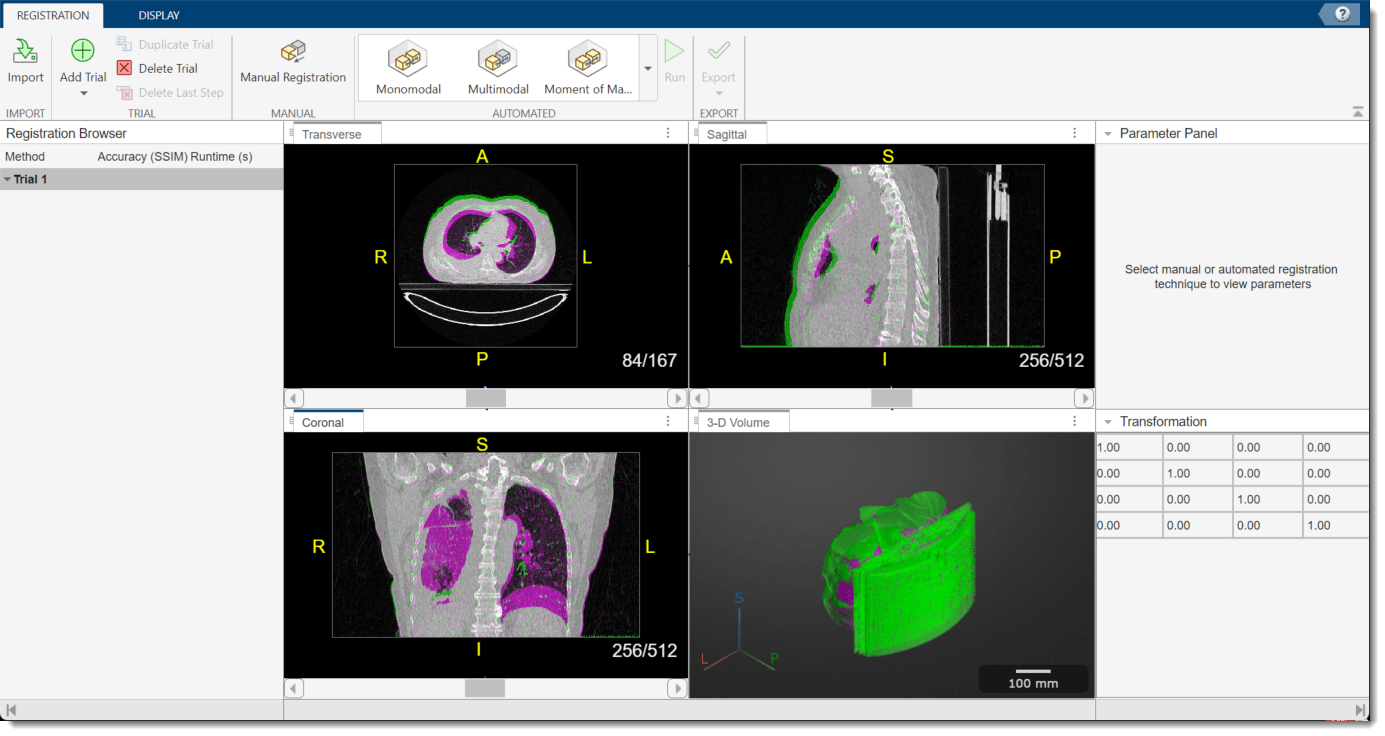
Apply Deformable Registration Technique
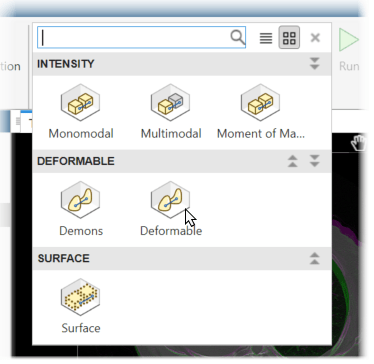
In this example, the volumes are already aligned, aside from the nonrigid motion due to breathing. To apply deformable registration, in the app toolstrip, in the Automated techniques gallery, select Deformable. This technique uses the same underlying algorithm as the imregdeform function.
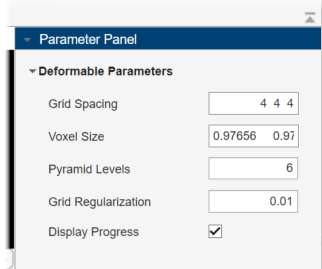
The Parameter Panel displays the adjustable parameters for the technique. By default, the app sets the Voxel Size value as [0.9766 0.9766 1.8], which matches the VoxelSpacing property of the medicalVolume object for the fixed volume. Specify the Pyramid Levels value as 6, and the Grid Regularization value as 0.01. Select Display Progress. These parameter values have been determined for this data set by using trial and error.
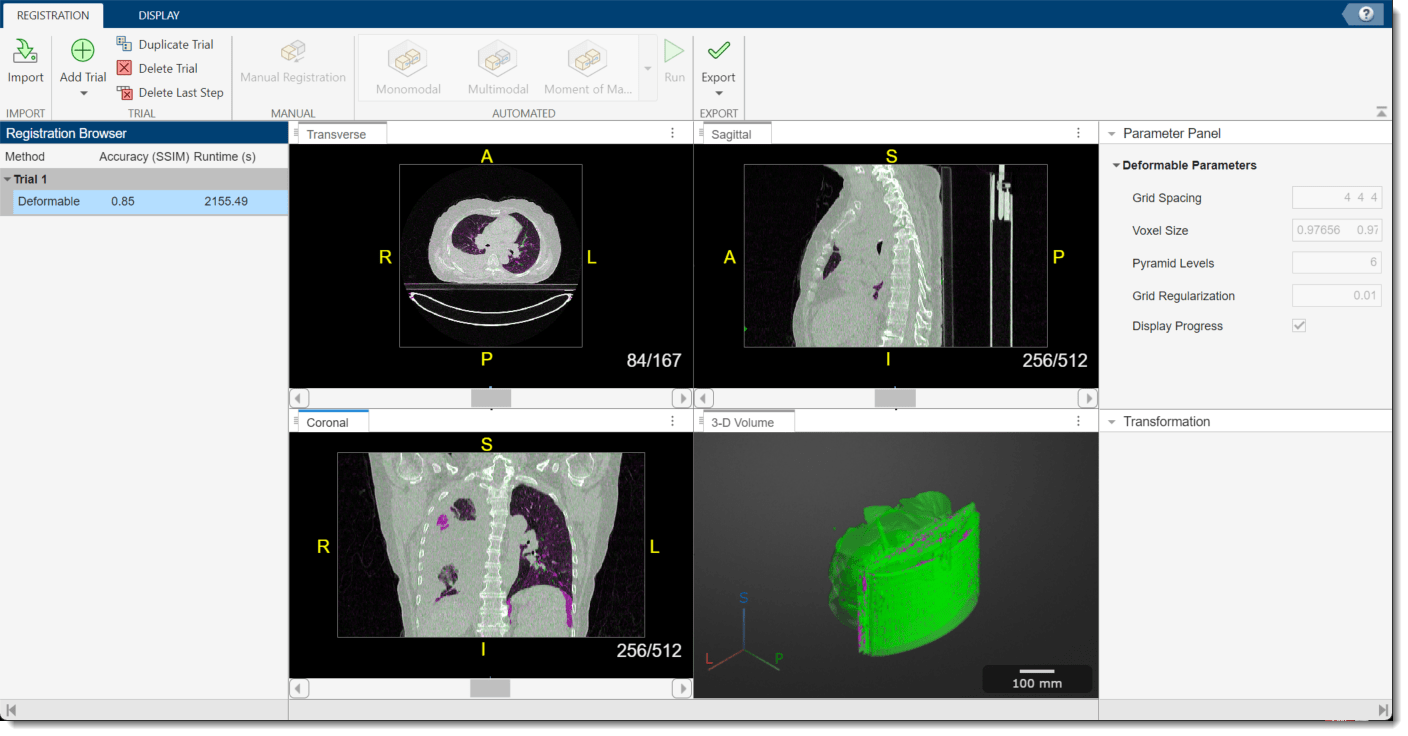
To run the registration technique, in the app toolstrip, select Run. The app runs the registration, which can take several minutes, and prints the progress in the MATLAB Command Window. When the app finishes, the 2-D and 3-D display panes update to show the registered volumes. When you run a deformable registration technique, the app clears the Transformation pane because the nonrigid displacement field is not described using a 4-by-4 matrix. You can assess the quality of the registration by visually checking the display panes and using the Accuracy (SSIM) metric in the Registration Browser pane, which provides the structural similarity index measure (SSIM) between the fixed and moving volumes.
Export Registration Results
To export the results, in the app toolstrip, select Export > To Workspace. In the dialog box, you can optionally change the name of the workspace variable to export. Click OK.
If you use the default name, the app creates a new workspace variable, movingReg, which is a structure containing these fields:
tCoarse— Contains the affine transformation object that describes any affine transformations applied in the trial, such as manual registration or a nondeformable automated technique. In this example, you apply only deformable registration, sotCoarsespecifies an identity transformation.dispField— Contains a 4-D array of size m-by-n-by-p-by-3 that describes the displacement field, where m, n, and p are the number of rows, columns, and slices of the registered volume, respectively. The first three dimensions correspond to row, column, and slice indices of the registered volume, and the fourth dimension contains the displacements in the x-, y-, and z-directions, in pixels. For example, the valuedispfield(1,1,1,:)returns a 3-element vector containing the x-, y-, and z-displacements at the voxel location (1, 1, 1).regVol— Contains the transformed, registered, moving volume as 3-D numeric array.
Examine Displacement Field
Create a new medicalVolume object that contains the registered image data and the spatial referencing information for the fixed exhale volume.
medVolReg = medicalVolume(movingReg.regVol,medVolExsp.VolumeGeometry);
Check the size of the displacement field. As expected, the displacement field is a 4-D array with the first three dimensions the same size as the volumes.
dispField = movingReg.dispField;
whos dispFieldName Size Bytes Class Attributes dispField 512x512x167x3 1050673152 double
Convert the displacements from pixel units to millimeters by multiplying each of the x-, y-, and z-displacements by the corresponding voxel spacing value.
dispFieldMM = dispField.*reshape(medVolInsp.VoxelSpacing,[1 1 1 3]);
Calculate the magnitude of the displacement by using the vecnorm function. Specify the p input as 2 to calculate the Euclidean distance, and the dimension along which to calculate the norm as 4.
dispMag = vecnorm(dispFieldMM,2,4);
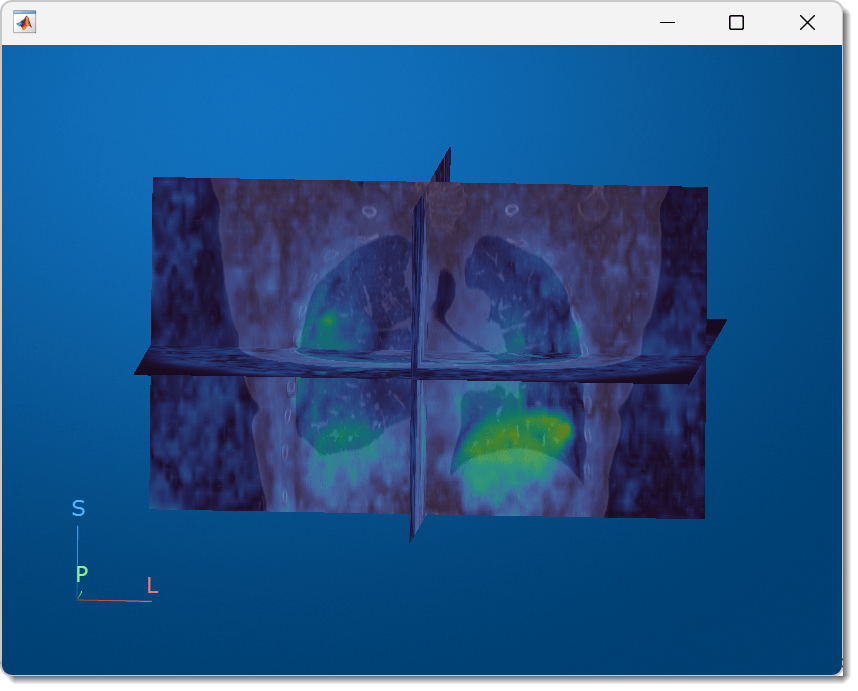
Display the displacement magnitude as an overlay on the original inhale CT volume. The cool heatmap colors indicate smaller displacements, and the warm heatmap regions indicate larger displacements. Consistent with breathing, the largest motion occurs within the bottom of the lungs, near the diaphragm. Regions outside the lungs have minimal motion, as the patient did not move or change position within the scanner other than to breathe.
volshow(medVolInsp, ... RenderingStyle="SlicePlanes", ... OverlayData=dispMag, ... OverlayDisplayRangeMode="data-range", ... OverlayColormap=turbo(256), ... OverlayAlphamap=0.5);

You can interact with the volume display to explore the displacement data, as shown in this animation. To view the coronal plane, click the P axis display indicator. Drag the cursor to rotate the volume slightly out of plane. To scroll through the coronal slices, pause on the coronal slice until it highlights red, then drag it along the anterior/posterior axis. To rotate the volume, click anywhere in the viewer and drag.
References
[1] Eslick, Enid M., John Kipritidis, Denis Gradinscak, Mark J. Stevens, Dale L. Bailey, Benjamin Harris, Jeremy T. Booth, and Paul J. Keall. “CT Ventilation as a Functional Imaging Modality for Lung Cancer Radiotherapy (CT-vs-PET-Ventilation-Imaging).” The Cancer Imaging Archive, 2022. https://doi.org/10.7937/3PPX-7S22.
[2] Eslick, Enid M., John Kipritidis, Denis Gradinscak, Mark J. Stevens, Dale L. Bailey, Benjamin Harris, Jeremy T. Booth, and Paul J. Keall. “CT Ventilation Imaging Derived from Breath Hold CT Exhibits Good Regional Accuracy with Galligas PET.” Radiotherapy and Oncology 127, no. 2 (May 2018): 267–73. https://doi.org/10.1016/j.radonc.2017.12.010.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository.” Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
See Also
Medical Registration
Estimator | imregdeform
