Examine Labeled Signal Set
Load into the MATLAB® Workspace the MAT file you created in the Label Signal Attributes, Regions of Interest, and Points example. Verify that the labeled signal set contains the definitions that you added using Signal Labeler.
load Whale_Songs
labelDefinitionsSummary(whalesongs)ans=3×9 table
LabelName LabelType LabelDataType Categories ValidationFunction DefaultValue Sublabels Tag Description
______________ ___________ _____________ ____________ __________________ ____________ ___________________________ ___ ____________________________
"WhaleType" "attribute" "categorical" {3×1 string} {["N/A" ]} {0×0 double} {0×0 double } "" "Whale type"
"MoanRegions" "roi" "logical" {["N/A" ]} {0×0 double} {0×0 double} {0×0 double } "" "Regions where moans occur"
"TrillRegions" "roi" "logical" {["N/A" ]} {0×0 double} {0×0 double} {1×1 signalLabelDefinition} "" "Regions where trills occur"
Verify that TrillPeaks is a sublabel of TrillRegions.
labelDefinitionsHierarchy(whalesongs)
ans =
'WhaleType
Sublabels: []
MoanRegions
Sublabels: []
TrillRegions
Sublabels: TrillPeaks
'
Retrieve the second member of the set. Retrieve the names of the timetable variables.
song = getSignal(whalesongs,2); summary(song)
song: 76579×1 timetable
Row Times:
Time: duration
Variables:
whale2: double
Statistics for applicable variables and row times:
NumMissing Min Median Max Mean Std
Time 0 0 sec 9.5722 sec 19.144 sec 9.5722 sec 5.5266 sec
whale2 0 -0.3733 0 0.3791 8.1118e-05 0.0392
Plot the signal.
t = song.Time; sng = song.whale2; plot(t,sng)

Visualize Labeled Regions
Use a signalMask object to display and identify the regions of interest that you labeled. For better display, change the label values from logical to categorical.
mvals = getLabelValues(whalesongs,2,'MoanRegions'); mvals.Value = categorical(repmat("moan",size(mvals,1),1)); tvals = getLabelValues(whalesongs,2,'TrillRegions'); tvals.Value = categorical(repmat("trill",size(tvals,1),1)); msk = signalMask([mvals;tvals],'SampleRate',1/seconds(t(2)-t(1))); plotsigroi(msk,sng)
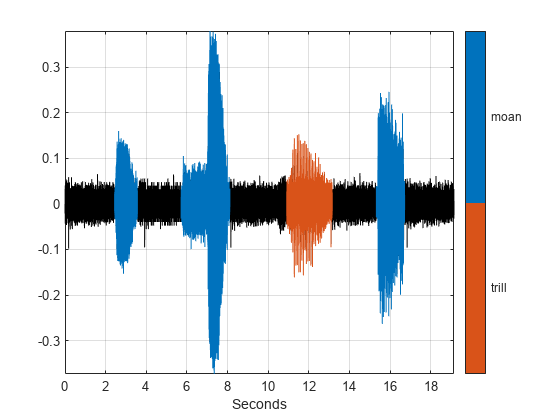
Visualize Labeled Points
Display and identify the trill peaks that you labeled.
pk = getLabelValues(whalesongs,2,{'TrillRegions','TrillPeaks'});
locs = zeros(size(pk,1),1);
for kj = 1:length(locs)
locs(kj) = find(seconds(t) == pk.Location(kj));
end
hold on
plot(seconds(t(locs)),sng(locs)+0.01,'v')
text(seconds(t(locs))+0.2,sng(locs)+0.05,int2str(cell2mat(pk.Value)))
hold off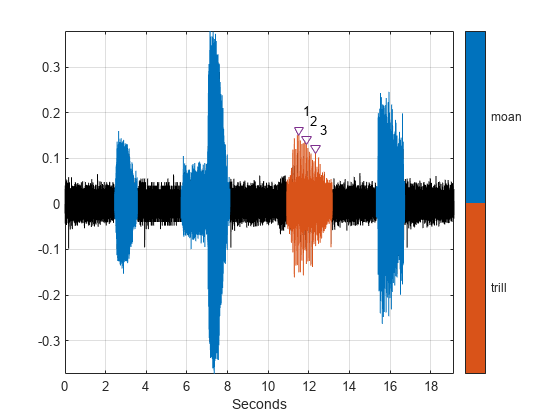
See Also
Apps
Objects
Topics
- Label Signal Attributes, Regions of Interest, and Points
- Label ECG Signals and Track Progress
- Automate Signal Labeling with Custom Functions
- Label Spoken Words in Audio Signals
- Use Signal Labeler App
- Import Data into Signal Labeler
- Create or Import Signal Label Definitions
- Label Signals Interactively or Automatically
- Custom Labeling Functions
- Customize Labeling View
- Dashboard
- Export Data and Create Data Sets
- Signal Labeler Usage Tips