SimBiology.gsa.MPGSA
Object containing multiparametric global sensitivity analysis (MPGSA) results
Description
Creation
Create a SimBiology.gsa.MPGSA object using sbiompgsa.
Properties
This property is read-only.
Expressions of model responses, specified as a cell array of character vectors.
Data Types: cell
This property is read-only.
Kolmogorov-Smirnov statistics, specified as a table. The table size is
[params,classifiers], where
params is the number of input parameters and
classifiers is the number of classifiers. Entry
[i,j] contains the
Kolmogorov-Smirnov statistic returned by kstest2 (Statistics and Machine Learning Toolbox) comparing the two eCDFs of the ith parameter
accepted and rejected by the jth classifier. If all samples are
accepted or rejected by the classifier, entry
[i,j] is set to
NaN.
The VariableNames property contains the
classifier expressions specified as the input to sbiompgsa. Long
expressions are truncated with the addition of '(classifier i)', where
i is the classifier index. The VariableDescriptions
property contains the untruncated classifier expressions.
Data Types: table
This property is read-only.
Computed eCDF data, specified as a cell array of numeric vectors. The size of the
array is
[params,4,classifiers], where
params is the number of input parameters and
classifiers is the number of classifiers.
Cells [i,1:2,j] contain the f (Statistics and Machine Learning Toolbox) and
x (Statistics and Machine Learning Toolbox) outputs from the ecdf (Statistics and Machine Learning Toolbox) function for the samples of parameter
i accepted by the classifier j.
Cells [i,3:4,j] contain the corresponding outputs for the samples
of parameter i rejected by the classifier
j.
If the classifier accepts all samples or rejects all samples, the corresponding eCDF data is empty.
Data Types: cell
This property is read-only.
Significance level of the two-sided Kolmogorov-Smirnov test, specified as a scalar
value in the range (0,1). For details, see kstest2 (Statistics and Machine Learning Toolbox).
Data Types: double
This property is read-only.
Asymptotic p-values for the Kolmogorov-Smirnov tests, specified as a table. The table size is [params,classifiers], where params is the number of input parameters and classifiers is the number of classifiers.
Entry [i,j] contains the
p-values returned by kstest2 (Statistics and Machine Learning Toolbox) comparing the two eCDFs of the
ith parameter being accepted and rejected by the
jth classifier. If all samples are accepted or rejected by the
classifier, entry [i,j] is set
to NaN.
The VariableNames property contains the
classifier expressions specified as the input to sbiompgsa. Long
expressions are truncated with the addition of '(classifier i)', where
i is the classifier index. The VariableDescriptions
property contains the untruncated classifier expressions.
Data Types: table
This property is read-only.
Flags indicating if the samples are accepted by the classifiers, specified as a table. The table size is [NumberSamples,classifiers], where NumberSamples is the number of parameter samples and classifiers is the number of classifiers.
The VariableNames property contains the
classifier expressions specified as the input to sbiompgsa. Long
expressions are truncated with the addition of '(classifier i)', where
i is the classifier index. The VariableDescriptions
property contains the untruncated classifier expressions.
Data Types: table
This property is read-only.
Names of model responses or observables, specified as a cell array of character vectors.
Data Types: char
This property is read-only.
Sampled parameter values, specified as a table. Each row represents one parameter set and each column represents one input parameter. For details, see Multiparametric Global Sensitivity Analysis (MPGSA).
Data Types: table
This property is read-only.
Simulation information, such as simulation data and parameter samples, used for multiparametric global sensitivity analysis, specified as a structure. The structure contains the following fields.
SimFunction—SimFunctionobject used for simulating model responses or observables.SimData—SimDataarray of size[NumberSamples,1], where NumberSamples is the number of samples. The array contains simulation results fromParameterSamples.OutputTimes— Numeric column vector containing the common time vector of allSimDataobjects.Bounds— Numeric matrix of size[params,2]. params is the number of input parameters. The first column contains the lower bounds and the second column contains the upper bounds for sensitivity inputs.This field is set to
[]if you provided parameter sample values as input when you calledsbiompgsa.DoseTables— Cell array of dose tables used for theSimFunctionevaluation.DoseTablesis the output ofgetTable(doseInput), where doseInput is the value specified for the'Doses'name-value pair argument in the call tosbiosobol,sbiompgsa, orsbioelementaryeffects. If no doses are applied, this field is set to[].ValidSample— Logical vector of size[NumberSamples,1]indicating whether a simulation for a particular sample failed. Resampling of the simulation data (SimData) can result inNaNvalues if the data is extrapolated. Such SimData are indicated as invalid.InterpolationMethod— Name of the interpolation method forSimData.SamplingMethod— Name of the sampling method used to drawParameterSamples. When you callsbiompgsawithsamples(sampled model quantities) as the input, this field of the corresponding results object is set to'unknown'.RandomState— Structure containing the state ofrngbefore drawingParameterSamples. When you callsbiompgsawithsamples(parameter sample values) as an input, this property of the corresponding results object is[].
Data Types: struct
Object Functions
plotData | Plot quantile summary of model simulations from global sensitivity analysis (requires Statistics and Machine Learning Toolbox) |
plot | Plot empirical CDF of multiparametric global sensitivity analysis |
bar | Create bar plot of multiparametric global sensitivity analysis statistics |
histogram | Plot histogram of multiparametric global sensitivity analysis results |
Examples
Load the target-mediated drug disposition (TMDD) model.
sbioloadproject tmdd_with_TO.sbprojGet the active configset and set the target occupancy (TO) as the response.
cs = getconfigset(m1);
cs.RuntimeOptions.StatesToLog = 'TO';Simulate the model and plot the TO profile.
sbioplot(sbiosimulate(m1,cs));
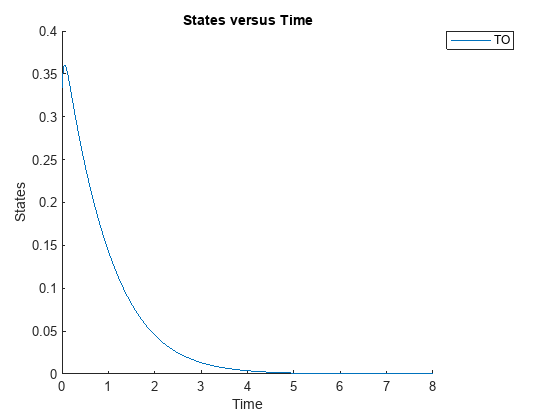
Define an exposure (area under the curve of the TO profile) threshold for the target occupancy.
classifier = 'trapz(time,TO) <= 0.1';Perform MPGSA to find sensitive parameters with respect to the TO. Vary the parameter values between predefined bounds to generate 10,000 parameter samples.
% Suppress an information warning that is issued during simulation. warnSettings = warning('off', 'SimBiology:sbservices:SB_DIMANALYSISNOTDONE_MATLABFCN_UCON'); rng(0,'twister'); % For reproducibility params = {'kel','ksyn','kdeg','km'}; bounds = [0.1, 1; 0.1, 1; 0.1, 1; 0.1, 1]; mpgsaResults = sbiompgsa(m1,params,classifier,Bounds=bounds,NumberSamples=10000)
mpgsaResults =
MPGSA with properties:
Classifiers: {'trapz(time,TO) <= 0.1'}
KolmogorovSmirnovStatistics: [4×1 table]
ECDFData: {4×4 cell}
SignificanceLevel: 0.0500
PValues: [4×1 table]
SupportHypothesis: [10000×1 table]
ParameterSamples: [10000×4 table]
Observables: {'TO'}
SimulationInfo: [1×1 struct]
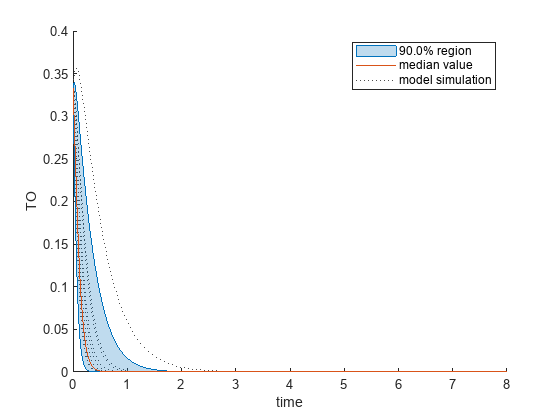
Plot the quantiles of the simulated model response.
plotData(mpgsaResults,ShowMedian=true,ShowMean=false);
Plot the empirical cumulative distribution functions (eCDFs) of the accepted and rejected samples. Except for km, none of the parameters shows a significant difference in the eCDFs for the accepted and rejected samples. The km plot shows a large Kolmogorov-Smirnov (K-S) distance between the eCDFs of the accepted and rejected samples. The K-S distance is the maximum absolute distance between two eCDFs curves.
h = plot(mpgsaResults);
% Resize the figure.
pos = h.Position(:);
h.Position(:) = [pos(1) pos(2) pos(3)*2 pos(4)*2];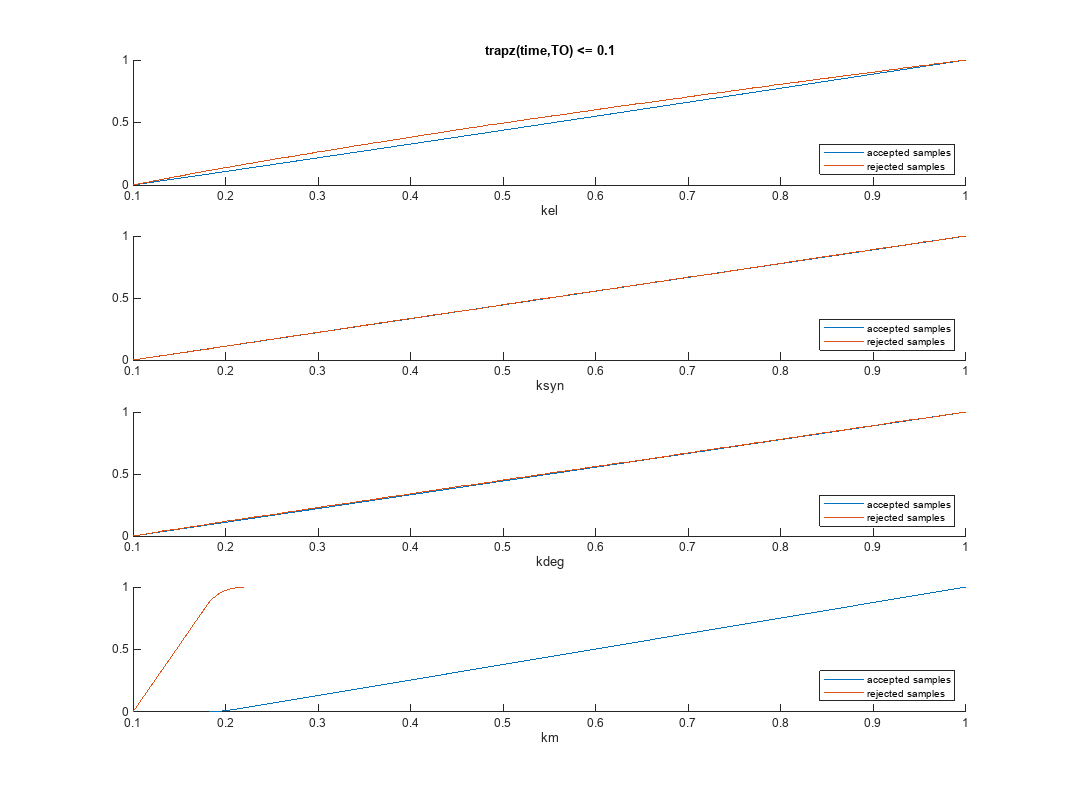
To compute the K-S distance between the two eCDFs, SimBiology uses a two-sided test based on the null hypothesis that the two distributions of accepted and rejected samples are equal. See kstest2 (Statistics and Machine Learning Toolbox) for details. If the K-S distance is large, then the two distributions are different, meaning that the classification of the samples is sensitive to variations in the input parameter. On the other hand, if the K-S distance is small, then variations in the input parameter do not affect the classification of samples. The results suggest that the classification is insensitive to the input parameter. To assess the significance of the K-S statistic rejecting the null-hypothesis, you can examine the p-values.
bar(mpgsaResults)
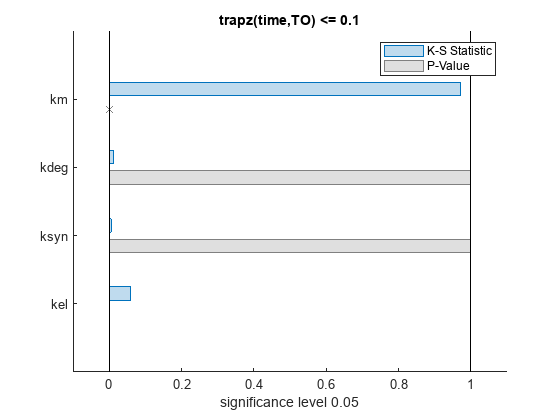
The bar plot shows two bars for each parameter: one for the K-S distance (K-S statistic) and another for the corresponding p-value. You reject the null hypothesis if the p-value is less than the significance level. A cross (x) is shown for any p-value that is almost 0. You can see the exact p-value corresponding to each parameter.
[mpgsaResults.ParameterSamples.Properties.VariableNames',mpgsaResults.PValues]
ans=4×2 table
Var1 trapz(time,TO) <= 0.1
________ _____________________
{'kel' } 0.0021877
{'ksyn'} 1
{'kdeg'} 0.99983
{'km' } 0
The p-values of km and kel are less than the significance level (0.05), supporting the alternative hypothesis that the accepted and rejected samples come from different distributions. In other words, the classification of the samples is sensitive to km and kel but not to other parameters (kdeg and ksyn).
You can also plot the histograms of accepted and rejected samples. The historgrams let you see trends in the accepted and rejected samples. In this example, the histogram of km shows that there are more accepted samples for larger km values, while the kel histogram shows that there are fewer rejected samples as kel increases.
h2 = histogram(mpgsaResults);
% Resize the figure.
pos = h2.Position(:);
h2.Position(:) = [pos(1) pos(2) pos(3)*2 pos(4)*2];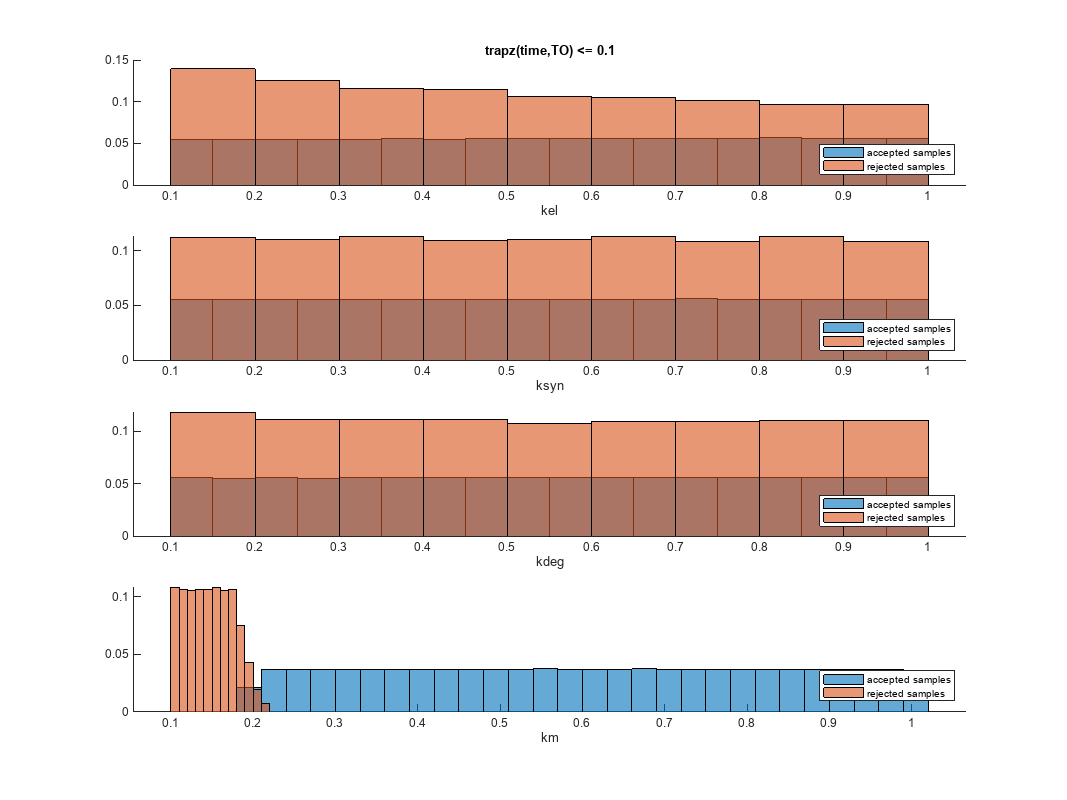
Restore the warning settings.
warning(warnSettings);
More About
Multiparametric global sensitivity analysis lets you study the relative
importance of parameters with respect to a classifier defined by model responses. A
classifier is an expression of model responses that evaluates to a logical vector.
sbiompgsa implements the MPSA method described by Tiemann et. al.
(see supporting information text S2) [1].
sbiompgsa performs the following steps.
Generate N parameter samples using a sampling method.
sbiompgsastores these samples as a table in a property,mpgsaResults.ParameterSamples, of the returned object. The number of rows is equal to the number of samples and the number of columns is equal to the number of input parameters.Tip
You can specify N and the sampling method using the
'NumberSamples'and'SamplingMethod'name-value pair arguments, respectively, when you callsbiompgsa.Calculate the model response by simulating the model for each parameter set, which is a single realization of the model parameter values. In this case, a parameter set is equal to a row in the
ParameterSamplestable.Evaluate the classifier. A classifier is an expression that evaluates to a logical vector. For instance, if your model response is the AUC of plasma drug concentration, you can define a classifier with a toxicity threshold of 0.8 where the AUC of the drug concentration above that threshold is considered toxic.
Parameter sets are then separated into two different groups, such as accepted (nontoxic) and rejected (toxic) groups.
For each input parameter, compute the empirical cumulative distribution functions (
ecdf(Statistics and Machine Learning Toolbox)) of accepted and rejected sample values.Compare the two eCDFs of accepted and rejected groups using the Two-Sample Kolmogorov-Smirnov Test (Statistics and Machine Learning Toolbox) to compute the Kolmogorov-Smirnov distance. The default significance level of the Kolmogorov-Smirnov test is
0.05. If two eCDFs are similar, the distance is small, meaning the model response is not sensitive with respect to the input parameter. If two eCDFs are different, the distance is large, meaning the model response is sensitive to the parameter.Note
The Kolmogorov-Smirnov test assumes that the samples follow a continuous distribution. Make sure that the eCDF plots are continuous as you increase the number of samples. If eCDFs are not continuous but step-like in the limit of infinite samples, then the results might not reflect the true sensitivities.
References
[1] Tiemann, Christian A., Joep Vanlier, Maaike H. Oosterveer, Albert K. Groen, Peter A. J. Hilbers, and Natal A. W. van Riel. “Parameter Trajectory Analysis to Identify Treatment Effects of Pharmacological Interventions.” Edited by Scott Markel. PLoS Computational Biology 9, no. 8 (August 1, 2013): e1003166. https://doi.org/10.1371/journal.pcbi.1003166.
Version History
Introduced in R2020a
See Also
sbiosobol | sbiompgsa | kstest2 (Statistics and Machine Learning Toolbox) | ecdf (Statistics and Machine Learning Toolbox) | Observable
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)