kfoldLoss
Classification loss for cross-validated kernel classification model
Description
loss = kfoldLoss(CVMdl)ClassificationPartitionedKernel) CVMdl. For every fold,
kfoldLoss computes the classification loss for validation-fold
observations using a model trained on training-fold observations.
By default, kfoldLoss returns the classification error.
loss = kfoldLoss(CVMdl,Name,Value)
Examples
Load the ionosphere data set. This data set has 34 predictors and 351 binary responses for radar returns, which are labeled either bad ('b') or good ('g').
load ionosphereCross-validate a binary kernel classification model using the data.
CVMdl = fitckernel(X,Y,'Crossval','on')
CVMdl =
ClassificationPartitionedKernel
CrossValidatedModel: 'Kernel'
ResponseName: 'Y'
NumObservations: 351
KFold: 10
Partition: [1×1 cvpartition]
ClassNames: {'b' 'g'}
ScoreTransform: 'none'
Properties, Methods
CVMdl is a ClassificationPartitionedKernel model. By default, the software implements 10-fold cross-validation. To specify a different number of folds, use the 'KFold' name-value pair argument instead of 'Crossval'.
Estimate the cross-validated classification loss. By default, the software computes the classification error.
loss = kfoldLoss(CVMdl)
loss = 0.0940
Alternatively, you can obtain the per-fold classification errors by specifying the name-value pair 'Mode','individual' in kfoldLoss.
Load the ionosphere data set. This data set has 34 predictors and 351 binary responses for radar returns, which are labeled either bad ('b') or good ('g').
load ionosphereCross-validate a binary kernel classification model using the data.
CVMdl = fitckernel(X,Y,'Crossval','on')
CVMdl =
ClassificationPartitionedKernel
CrossValidatedModel: 'Kernel'
ResponseName: 'Y'
NumObservations: 351
KFold: 10
Partition: [1×1 cvpartition]
ClassNames: {'b' 'g'}
ScoreTransform: 'none'
Properties, Methods
CVMdl is a ClassificationPartitionedKernel model. By default, the software implements 10-fold cross-validation. To specify a different number of folds, use the 'KFold' name-value pair argument instead of 'Crossval'.
Create an anonymous function that measures linear loss, that is,
is the weight for observation j, is the response j (–1 for the negative class and 1 otherwise), and is the raw classification score of observation j.
linearloss = @(C,S,W,Cost)sum(-W.*sum(S.*C,2))/sum(W);
Custom loss functions must be written in a particular form. For rules on writing a custom loss function, see the 'LossFun' name-value pair argument.
Estimate the cross-validated classification loss using the linear loss function.
loss = kfoldLoss(CVMdl,'LossFun',linearloss)loss = -0.7792
Input Arguments
Cross-validated, binary kernel classification model, specified as a ClassificationPartitionedKernel model object. You can create a
ClassificationPartitionedKernel model by using fitckernel
and specifying any one of the cross-validation name-value pair arguments.
To obtain estimates, kfoldLoss applies the same data used to
cross-validate the kernel classification model (X and
Y).
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: kfoldLoss(CVMdl,'Folds',[1 3 5]) specifies to use only the
first, third, and fifth folds to calculate the classification loss.
Fold indices for prediction, specified as the comma-separated pair consisting of
'Folds' and a numeric vector of positive integers. The elements
of Folds must be within the range from 1 to
CVMdl.KFold.
The software uses only the folds specified in Folds for
prediction.
Example: 'Folds',[1 4 10]
Data Types: single | double
Loss function, specified as the comma-separated pair consisting of
'LossFun' and a built-in loss function name or a function handle.
This table lists the available loss functions. Specify one using its corresponding value.
Value Description "binodeviance"Binomial deviance "classifcost"Observed misclassification cost "classiferror"Misclassified rate in decimal "exponential"Exponential loss "hinge"Hinge loss "logit"Logistic loss "mincost"Minimal expected misclassification cost (for classification scores that are posterior probabilities) "quadratic"Quadratic loss 'mincost'is appropriate for classification scores that are posterior probabilities. For kernel classification models, logistic regression learners return posterior probabilities as classification scores by default, but SVM learners do not (seekfoldPredict).Specify your own function by using function handle notation.
Assume that
nis the number of observations inX, andKis the number of distinct classes (numel(CVMdl.ClassNames), whereCVMdlis the input model). Your function must have this signature:lossvalue =lossfun(C,S,W,Cost)The output argument
lossvalueis a scalar.You specify the function name (
lossfun).Cis ann-by-Klogical matrix with rows indicating the class to which the corresponding observation belongs. The column order corresponds to the class order inCVMdl.ClassNames.Construct
Cby settingC(p,q) = 1, if observationpis in classq, for each row. Set all other elements of rowpto0.Sis ann-by-Knumeric matrix of classification scores. The column order corresponds to the class order inCVMdl.ClassNames.Sis a matrix of classification scores, similar to the output ofkfoldPredict.Wis ann-by-1 numeric vector of observation weights. If you passW, the software normalizes the weights to sum to1.Costis aK-by-Knumeric matrix of misclassification costs. For example,Cost = ones(K) – eye(K)specifies a cost of0for correct classification, and1for misclassification.
Example: 'LossFun',@lossfun
Data Types: char | string | function_handle
Aggregation level for the output, specified as the comma-separated pair consisting of
'Mode' and 'average' or
'individual'.
This table describes the values.
| Value | Description |
|---|---|
'average' | The output is a scalar average over all folds. |
'individual' | The output is a vector of length k containing one value per fold, where k is the number of folds. |
Example: 'Mode','individual'
Output Arguments
Classification loss, returned as a numeric scalar or numeric column vector.
If Mode is 'average', then
loss is the average classification loss over all folds.
Otherwise, loss is a k-by-1 numeric column
vector containing the classification loss for each fold, where k is
the number of folds.
More About
Classification loss functions measure the predictive inaccuracy of classification models. When you compare the same type of loss among many models, a lower loss indicates a better predictive model.
Consider the following scenario.
L is the weighted average classification loss.
n is the sample size.
yj is the observed class label. The software codes it as –1 or 1, indicating the negative or positive class (or the first or second class in the
ClassNamesproperty), respectively.f(Xj) is the positive-class classification score for observation (row) j of the predictor data X.
mj = yjf(Xj) is the classification score for classifying observation j into the class corresponding to yj. Positive values of mj indicate correct classification and do not contribute much to the average loss. Negative values of mj indicate incorrect classification and contribute significantly to the average loss.
The weight for observation j is wj. The software normalizes the observation weights so that they sum to the corresponding prior class probability stored in the
Priorproperty. Therefore,
Given this scenario, the following table describes the supported loss functions that you can specify by using the LossFun name-value argument.
| Loss Function | Value of LossFun | Equation |
|---|---|---|
| Binomial deviance | "binodeviance" | |
| Observed misclassification cost | "classifcost" | where is the class label corresponding to the class with the maximal score, and is the user-specified cost of classifying an observation into class when its true class is yj. |
| Misclassified rate in decimal | "classiferror" | where I{·} is the indicator function. |
| Cross-entropy loss | "crossentropy" |
The weighted cross-entropy loss is where the weights are normalized to sum to n instead of 1. |
| Exponential loss | "exponential" | |
| Hinge loss | "hinge" | |
| Logistic loss | "logit" | |
| Minimal expected misclassification cost | "mincost" |
The software computes the weighted minimal expected classification cost using this procedure for observations j = 1,...,n.
The weighted average of the minimal expected misclassification cost loss is |
| Quadratic loss | "quadratic" |
If you use the default cost matrix (whose element value is 0 for correct classification
and 1 for incorrect classification), then the loss values for
"classifcost", "classiferror", and
"mincost" are identical. For a model with a nondefault cost matrix,
the "classifcost" loss is equivalent to the "mincost"
loss most of the time. These losses can be different if prediction into the class with
maximal posterior probability is different from prediction into the class with minimal
expected cost. Note that "mincost" is appropriate only if classification
scores are posterior probabilities.
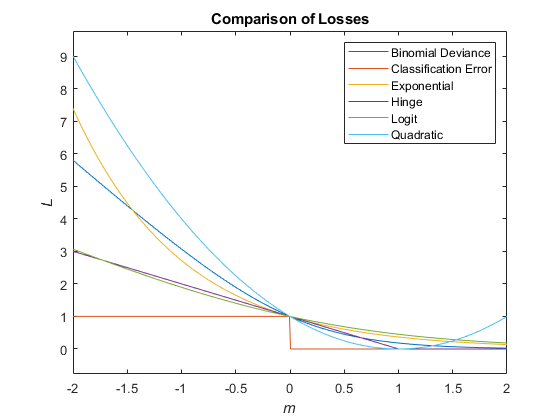
This figure compares the loss functions (except "classifcost",
"crossentropy", and "mincost") over the score
m for one observation. Some functions are normalized to pass through
the point (0,1).
Extended Capabilities
This function fully supports GPU arrays. For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2018bkfoldLoss fully supports GPU arrays.
Starting in R2023b, the following classification model object functions use observations with missing predictor values as part of resubstitution ("resub") and cross-validation ("kfold") computations for classification edges, losses, margins, and predictions.
In previous releases, the software omitted observations with missing predictor values from the resubstitution and cross-validation computations.
If you specify a nondefault cost matrix when you train the input model object, the kfoldLoss function returns a different value compared to previous releases.
The kfoldLoss function uses the
observation weights stored in the W property. Also, the function uses the
cost matrix stored in the Cost property if you specify the
LossFun name-value argument as "classifcost" or
"mincost". The way the function uses the W and
Cost property values has not changed. However, the property values stored in the input model object have changed for a
model with a nondefault cost matrix, so the function might return a different value.
For details about the property value change, see Cost property stores the user-specified cost matrix.
If you want the software to handle the cost matrix, prior
probabilities, and observation weights in the same way as in previous releases, adjust the prior
probabilities and observation weights for the nondefault cost matrix, as described in Adjust Prior Probabilities and Observation Weights for Misclassification Cost Matrix. Then, when you train a
classification model, specify the adjusted prior probabilities and observation weights by using
the Prior and Weights name-value arguments, respectively,
and use the default cost matrix.
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)