makima
修正 Akima 分段三次 Hermite 插值
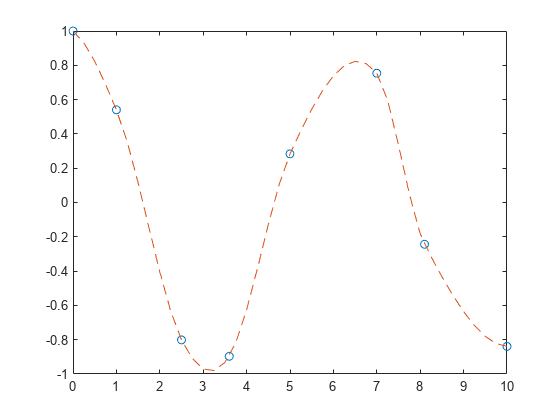
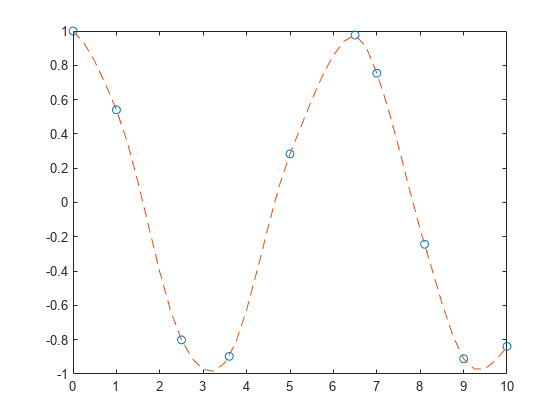
说明
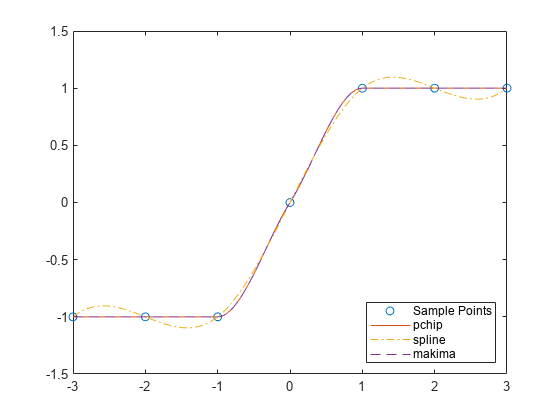
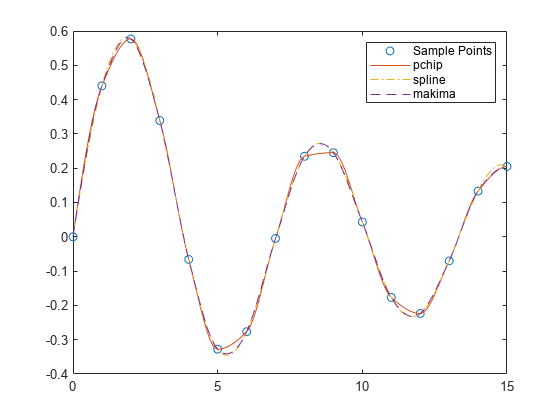
示例
输入参数
输出参量
详细信息
参考
[1] Akima, Hiroshi. "A new method of interpolation and smooth curve fitting based on local procedures." Journal of the ACM (JACM) , 17.4, 1970, pp. 589–602.
[2] Akima, Hiroshi. "A method of bivariate interpolation and smooth surface fitting based on local procedures." Communications of the ACM , 17.1, 1974, pp. 18–20.
扩展功能
版本历史记录
在 R2019b 中推出