copulafit
Fit copula to data
Syntax
Description
___ = copulafit(___, returns
any of the previous syntaxes, with additional options specified by
one or more Name,Value)Name,Value pair arguments. For example,
you can specify the confidence interval to compute, or specify control
parameters for the iterative parameter estimation algorithm using
a options structure.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Algorithms
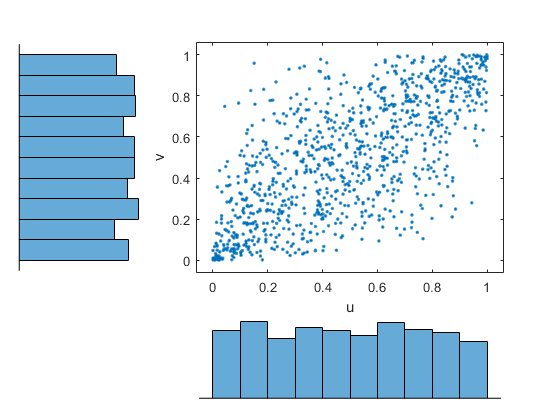
By default, copulafit uses maximum likelihood
to fit a copula to u. When u contains
data transformed to the unit hypercube by parametric estimates of
their marginal cumulative distribution functions, this is known as
the Inference Functions for Margins (IFM) method.
When u contains data transformed by the empirical
cdf (see ecdf), this is known
as Canonical Maximum Likelihood (CML).
References
[1] Bouyé, E., V. Durrleman, A. Nikeghbali, G. Riboulet, and T. Roncalli. “Copulas for Finance: A Reading Guide and Some Applications.” Working Paper. Groupe de Recherche Opérationnelle, Crédit Lyonnais, Paris, 2000.
Version History
Introduced in R2007b