tune
Tune Bayesian nonlinear non-Gaussian state-space model posterior sampler
Since R2024b
Syntax
Description
The Markov chain Monte Carlo (MCMC) method used by the simulate function
to sample from the posterior distribution requires a carefully specified proposal
distribution. You can use the tune function to search for an
adequate posterior mode for simulate, to improve
the proposal distribution, and therefore, the acceptance rate of the proposed posterior
draws.
[
returns optimized proposal distribution parameter mean vector params,Proposal] = tune(PriorMdl,Y,params0)params
and scale matrix Proposal to improve the MCMC sampler used by simulate.
PriorMdl is the Bayesian nonlinear non-Gaussian state-space model
that specifies the state-space model structure (likelihood) and prior distribution,
Y is the data for the likelihood, and params0 is
the vector of initial values for the unknown state-space model parameters Θ in
PriorMdl.
[
specifies additional options using one or more name-value arguments. For example,
params,Proposal] = tune(PriorMdl,Y,params0,Name=Value)tune(PriorMdl,Y,params0,Hessian="opg",Display="off") uses the
outer-product of gradients method to compute the Hessian matrix and suppresses the display
of the optimized values.
Examples
Simulate observed responses from a known nonlinear state-space model, then treat the model as Bayesian and draw parameters from the posterior distribution. Tune the proposal distribution of the MCMC sampler by using tune.
Consider this data-generating process (DGP).
where the series is a standard Gaussian series of random variables.
Simulate a series of 200 observations from the process.
rng(1,"twister") % For reproducibility T = 200; thetaDGP = [0.7; 0.2; 3]; numparams = numel(thetaDGP); MdlXSim = arima(AR=thetaDGP(1),Variance=thetaDGP(2), ... Constant=0); xsim = simulate(MdlXSim,T); y = random("poisson",thetaDGP(3)*exp(xsim)); figure plot(y)
Suppose the structure of the DGP is known, but the model parameters trueTheta are unknown, explicitly
Consider a Bayesian nonlinear non-Gaussian state-space model representing the unknown parameters. Assume that the prior distributions are:
, that is, a truncated normal distribution with .
, that is, an inverse gamma distribution with shape and scale .
, that is, a gamma distribution with shape and scale .
The Local Functions section contains the functions paramMap and priorDistribution required to specify the Bayesian nonlinear state-space model. The paramMap function specifies the state-space model structure and initial state moments. The priorDistribution function returns the log of the joint prior distribution of the state-space model parameters. You can use the functions only within this script.
Create a Bayesian nonlinear state-space model for the DGP.
Arbitrarily choose values for the hyperparameters.
Indicate that the state-space model observation equation is expressed as a distribution.
To speed up computations, the arguments
AandLogYof theparamMapfunction are written to enable simultaneous evaluation of the transition and observation densities of multiple particles. Specify this characteristic by using theMultipointname-value argument.
% pi(phi,sigma2) hyperparameters m0 = 0; v02 = 1; a0 = 1; b0 = 1; % pi(lambda) hyperparameters alpha0 = 3; beta0 = 1; hyperparams = [m0 v02 a0 b0 alpha0 beta0]; PriorMdl = bnlssm(@paramMap,@(x)priorDistribution(x,hyperparams), ... ObservationForm="distribution",Multipoint=["A" "LogY"]);
PriorMdl is a bnlssm model specifying the state-space model structure and prior distribution of the state-space model parameters. Because PriorMdl contains unknown values, it serves as a template for posterior estimation with observations.
The simulate function requires a proposal distribution scale matrix. You can obtain a data-driven proposal scale matrix by using the tune function. Alternatively, you can supply your own scale matrix.
Obtain a data-driven scale matrix by using the tune function. Supply an arbitrarily chosen set of initial parameter values.
theta0 = [0.5; 0.1; 2]; [theta0,Proposal] = tune(PriorMdl,y,theta0)
Optimization and Tuning
| Params0 Optimized ProposalStd
----------------------------------------
c(1) | 0.5000 0.6081 0.0922
c(2) | 0.1000 0.2190 0.0503
c(3) | 2 2.8497 0.2918
theta0 = 3×1
0.6081
0.2190
2.8497
Proposal = 3×3
0.0085 -0.0026 0.0033
-0.0026 0.0025 -0.0040
0.0033 -0.0040 0.0852
theta0 is a 3-by-1 estimate of the posterior mode and Proposal is the corresponding Hessian matrix. Both outputs are the optimized moments of the proposal distribution, the latter of which is up to a proportionality constant. tune displays convergence information and an estimation table, which you can suppress by using the Display options of the optimizer and tune.
Draw 1000 random parameter vectors from the posterior distribution. Specify the simulated response path as observed responses and the optimized values returned by tune for the initial parameter values and the proposal distribution.
[Theta,accept] = simulate(PriorMdl,y,theta0,Proposal); accept
accept = 0.4210
Theta is a 3-by-1000 matrix of randomly drawn parameters from the posterior distribution. Rows correspond to the elements of the input argument theta of the functions paramMap and priorDistribution.
accept is the proposal acceptance probability. In this case, simulate accepts 42% of the proposal draws.
Plot trace plots of the posterior sample.
paramnames = ["\phi" "\sigma^2" "\lambda"]; figure h = tiledlayout(3,1); for j = 1:numparams nexttile plot(Theta(j,:)) hold on yline(thetaDGP(j)) ylabel(paramnames(j)) end title(h,"Posterior Trace Plots")
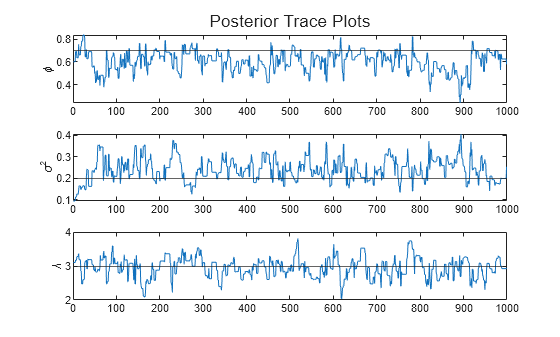
The sampler eventually settles at near the true values of the parameters. In this case, the sample shows serial correlation and transient behavior. You can remedy serial correlation in the sample by adjusting the Thin name-value argument, and you can remedy transient effects by increasing the burn-in period using the BurnIn name-value argument.
Local Functions
These functions specify the state-space model parameter mappings, in distribution form, and the log prior distribution of the parameters.
function [A,B,LogY,Mean0,Cov0,StateType] = paramMap(theta) A = theta(1); B = sqrt(theta(2)); LogY = @(y,x)y.*(x + log(theta(3)))- exp(x).*theta(3); Mean0 = 0; Cov0 = 2; StateType = 0; % Stationary state process end function logprior = priorDistribution(theta,hyperparams) % Prior of phi m0 = hyperparams(1); v20 = hyperparams(2); pphi = makedist("normal",mu=m0,sigma=sqrt(v20)); pphi = truncate(pphi,-1,1); lpphi = log(pdf(pphi,theta(1))); % Prior of sigma2 a0 = hyperparams(3); b0 = hyperparams(4); lpsigma2 = -a0*log(b0) - log(gamma(a0)) + (-a0-1)*log(theta(2)) - ... 1./(b0*theta(2)); % Prior of lambda alpha0 = hyperparams(5); beta0 = hyperparams(6); plambda = makedist("gamma",alpha0,beta0); lplambda = log(pdf(plambda,theta(3))); logprior = lpphi + lpsigma2 + lplambda; end
Adjust the tuning algorithm that obtains optimal proposal moments for the MCMC sampler used to draw posterior samples from a Bayesian nonlinear stochastic volatility model. The response series is daily S&P 500 closing returns. For a full exposition of this problem, see Fit Bayesian Stochastic Volatility Model to S&P 500 Volatility.
Load the data set Data_GlobalIdx1.mat, and then extract the S&P 500 closing prices (last column of the matrix Data).
load Data_GlobalIdx1 sp500 = Data(:,end); T = numel(sp500); dts = datetime(dates,ConvertFrom="datenum");
Convert the price series to returns, and then center the returns.
retsp500 = price2ret(sp500); y = retsp500 - mean(retsp500); retdts = dts(2:end);
Consider the nonlinear state-space form of the stochastic volatility model for observed asset returns :
where:
for daily returns.
is a latent AR(1) process representing the conditional volatility series.
are are mutually independent and individually iid random standard Gaussian noise series.
The linear state-space coefficient matrices are:
The observation equation is nonlinear. Because is a standard Gaussian random variable, the conditional distribution of given and the parameters is Gaussian with a mean of 0 and standard deviation .
The local function paramMap, which uses the distribution form for the observation equation, specifies this model structure. Unlike the parameter mapping function in Fit Bayesian Stochastic Volatility Model to S&P 500 Volatility, paramMap reduces the number of states to one for computational efficiency.
Assume a flat prior, that is, the log prior is 0 over the support of the parameters and -Inf everywhere else. The local function flatLogPrior specifies the prior.
Create a Bayesian nonlinear state-space model that specifies the model structure and prior distribution. Specify that the observation equation is in distribution form and that bnlssm functions can evaluate multiple particles simultaneously for A and LogY.
PriorMdl = bnlssm(@paramMap,@flatLogPrior,ObservationForm="distribution", ... Multipoint=["A" "LogY"]);
Tune the proposal distribution. Specify an arbitrarily chosen set of initial parameter values and the following settings:
Set the search interval for to (-1,1). Specified search intervals apply only to tuning the proposal.
Set the search interval for to (0,).
Specify computing the Hessian by the outer product of gradients.
For computational speed, do not sort the particles by using Hilbert sorting.
theta0 = [0.2 0 0.5]; lower = [-1; -Inf; 0]; upper = [1; Inf; Inf]; rng(1) [params,Proposal] = tune(PriorMdl,y,theta0,Lower=lower,Upper=upper, ... NumParticles=500,Hessian="opg",SortParticles=false);
Optimization and Tuning
| Params0 Optimized ProposalStd
----------------------------------------
c(1) | 0.2000 0.9949 0.0030
c(2) | 0 -0.0155 0.0116
c(3) | 0.5000 0.1495 0.0130
Obtain a sample from the posterior distribution. Specify the optimal proposal a burn-in period of 1e4 and retain every 25th draw to reduce serial correlation among the draws. The sampler, with these settings, takes some time to complete.
burnin = 1e4;
thin = 25;
[ThetaPost,accept] = simulate(PriorMdl,y,params,Proposal, ...
BurnIn=burnin,Thin=thin);
acceptaccept = 0.5131
Determine the quality of the posterior sample by plotting trace plots of the parameter draws.
figure
plot(ThetaPost(1,:))
title("Trace Plot: \beta")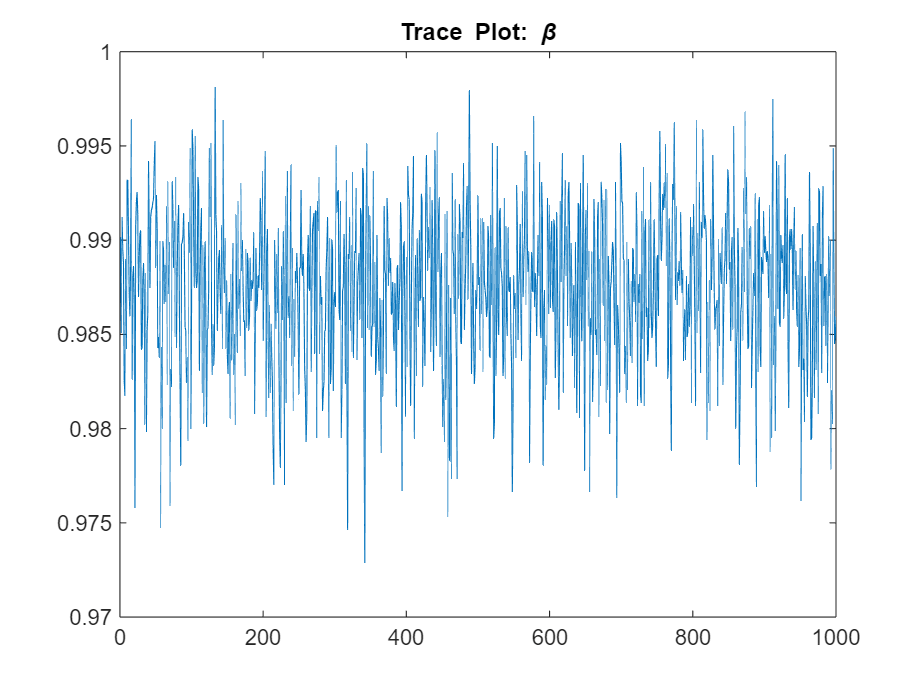
figure
plot(ThetaPost(2,:))
title("Trace Plot: \alpha")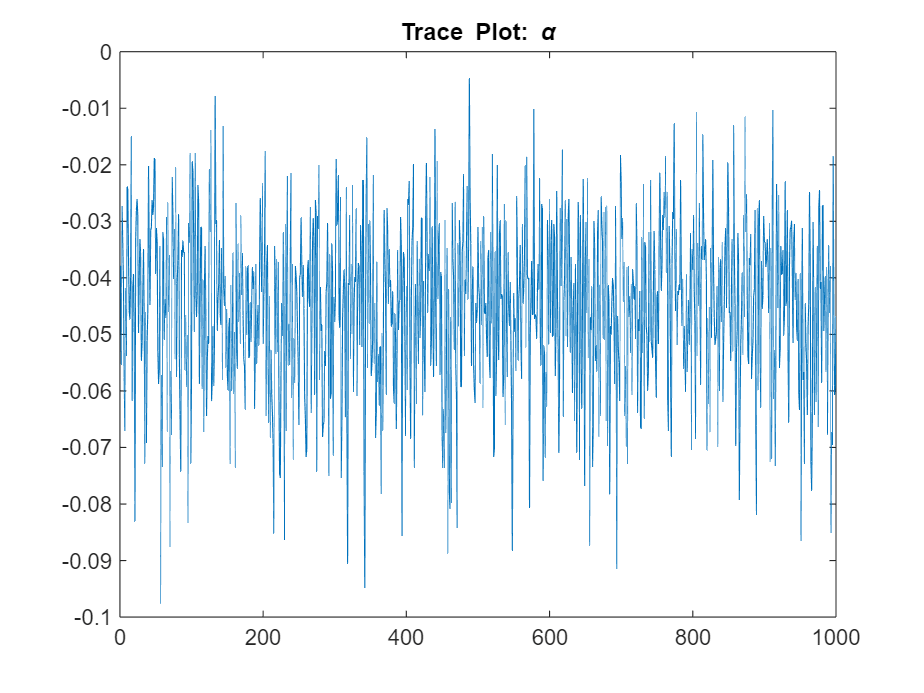
figure
plot(ThetaPost(3,:))
title("Trace Plot: \sigma")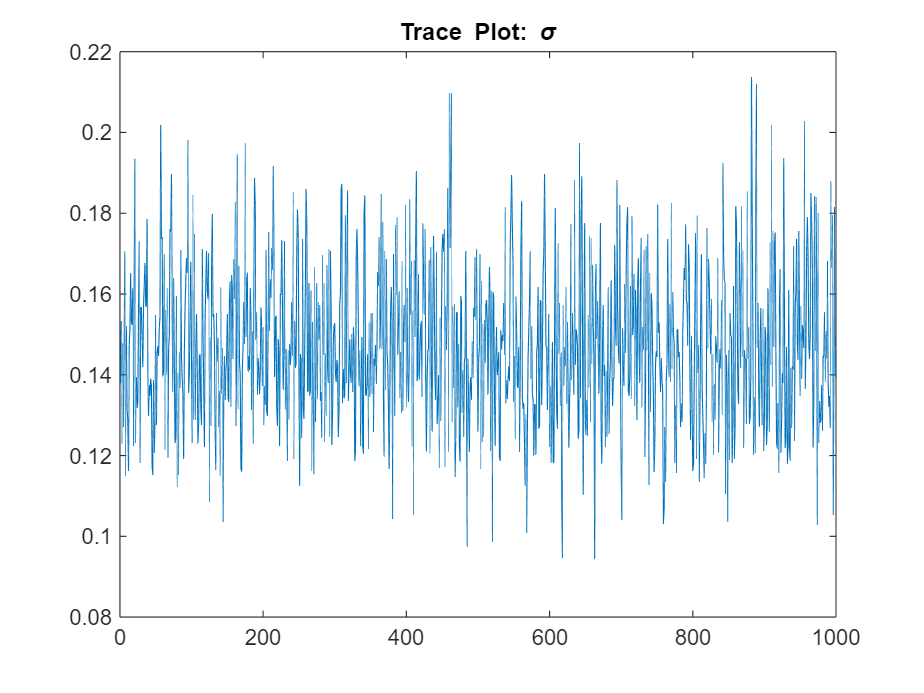
The posterior samples show good mixing with some minor serial correlation.
Local Functions
This example uses the following functions. paramMap is the parameter-to-matrix mapping function and flatLogPrior is the log prior distribution of the parameters.
function [A,B,LogY,Mean0,Cov0,StateType] = paramMap(theta) A = @(x) theta(2) + theta(1).*x; B = theta(3); LogY = @(y,x)-0.5.*x-0.5*365*y*y.*exp(-x); Mean0 = theta(2)./(1 - theta(1)); Cov0 = (theta(3)^2)./(1 - theta(1)^2); StateType = 0; end function logprior = flatLogPrior(theta) % flatLogPrior computes the log of the flat prior density for the three % variables in theta. Log probabilities for parameters outside the % parameter space are -Inf. paramcon = zeros(numel(theta),1); % theta(1) is the lag 1 term in a stationary AR(1) model. The % value needs to be within the unit circle. paramcon(1) = abs(theta(1)) >= 1 - eps; % alpha must be finite paramcon(2) = ~isfinite(theta(2)); % Standard deviation of the state disturbance theta(3) must be positive. paramcon(3) = theta(3) <= eps; if sum(paramcon) > 0 logprior = -Inf; else logprior = 0; % Prior density is proportional to 1 for all values % in the parameter space. end end
This example compares the performance of several SMC proposal particle resampling methods for obtaining random paths of parameter values from the posterior distribution of a quasi-nonnegative constrained state space model, which models nonnegative state quantities such as interest rates and prices:
and are iid standard Gaussian random variables.
Generate Artificial Data
Consider the following data-generating process (DGP)
Generate a random series of 200 observations from the DGP.
a0 = 0.1; a1 = 0.95; b = 1; d = 0.5; theta = [a0; a1; b; d]; T = 200; % Preallocate variables x = zeros(T,1); y = zeros(T,1); rng(0,"twister") % For reproducibility u = randn(T,1); e = randn(T,1); for t = 2:T x(t) = max(0,a0 + a1*x(t-1)) + b*u(t); y(t) = x(t) + d*e(t); end
Create Bayesian Nonlinear State-Space Model
The Local Functions section contains two functions required to specify the Bayesian nonlinear state-space model: the state-space model parameter mapping function paramMap and the prior distribution of the parameters priorDistribution. You can use the functions only within this script.
The paramMap function has these qualities:
Functions can simultaneously evaluate the state equation for multiple values of
A. Therefore, you can speed up calculations by specifying theMultipoint="A"option.The observation equation is in equation form, that is, the function composing the states is nonlinear and the innovation series is additive, linear, and Gaussian.
The priorDistribution function specifies a flat prior, which has a density that is proportional to 1 everywhere in the parameter space; it constrains the error standard deviations to be positive.
Create a Bayesian nonlinear state-space model characterized by the system.
PriorMdl = bnlssm(@paramMap,@priorDistribution,Multipoint="A");Draw Random Parameter Paths from Posterior Using Each Proposal Sampler Method
Obtain random parameter paths from the joint posterior distribution using the bootstrap, optimal, and one-pass unscented particle resampling methods. Tune the sampler initialized from a randomly drawn set of values in [0,1], thin the sample by a factor of 10, specify a burn-in period of 100, and draw 1000 paths.
numDraws = 1000; smcMethod = ["bootstrap" "optimal" "unscented"]; for j = 3:-1:1 % Set last element first to preallocate entire cell vector params0 = rand(4,1); [params,Proposal] = tune(PriorMdl,y,params0,Display="off",NewSamples=smcMethod(j)); thetaSim{j} = simulate(PriorMdl,y,params,Proposal,NewSamples=smcMethod(j), ... NumDraws=numDraws,Thin=10,Burnin=100); end
Separately for each parameter, draw histograms of the posterior draws for each method on the same plot.
figure tiledlayout(2,2) varnames = ["a0" "a1" "b" "d"]; for j = 1:numel(theta) nexttile h1 = histogram(thetaSim{1}(j,:)); hold on h2 = histogram(thetaSim{2}(j,:)); h3 = histogram(thetaSim{3}(j,:)); h4 = xline(theta(j),"--",LineWidth=2); hold off title(varnames(j)) end legend([h1 h2 h3 h4],[smcMethod "True"])
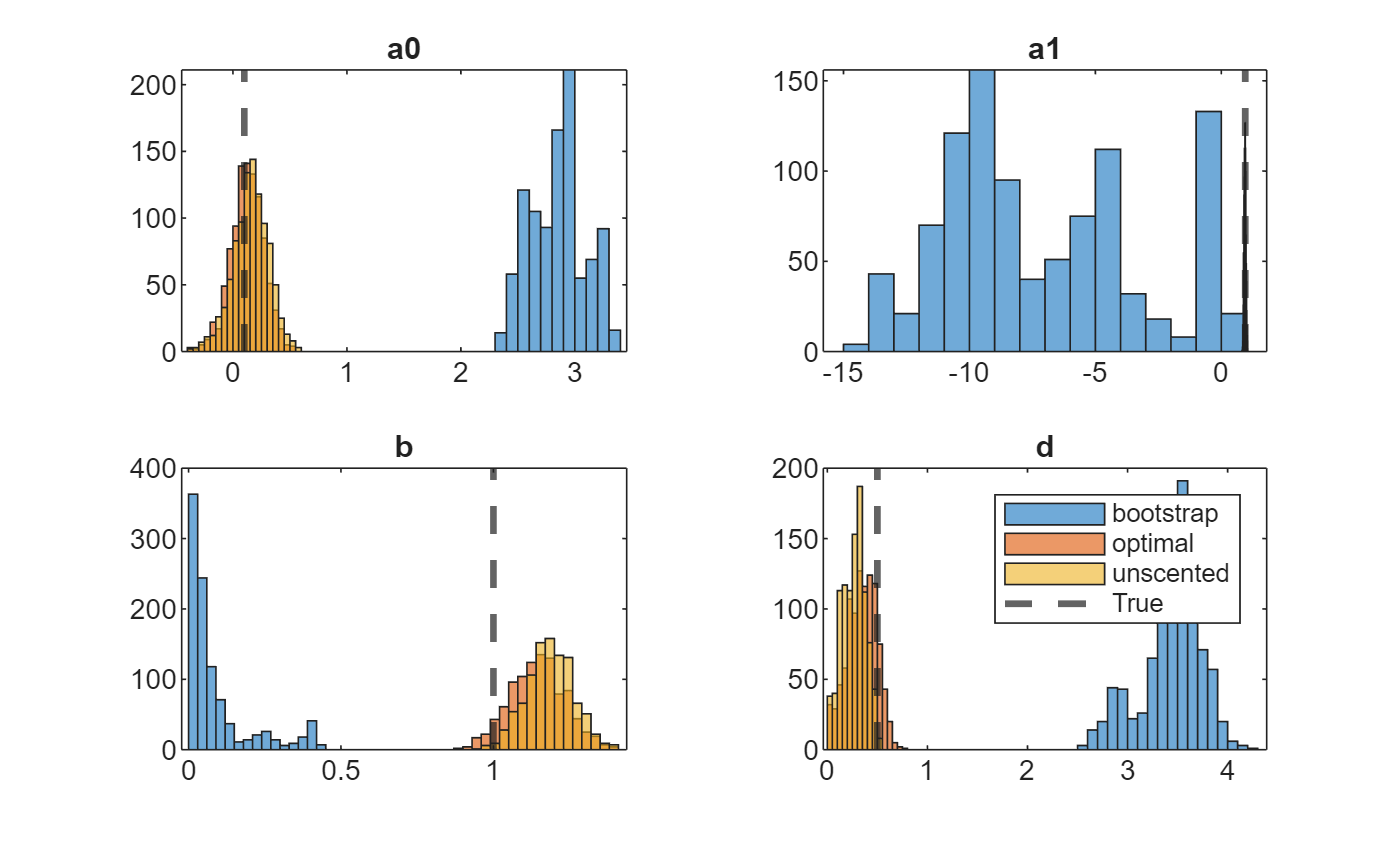
In this case, the optimal and one-pass unscented methods yield distributions closer to the true values than the bootstrap method.
Local Functions
These functions specify the state-space model parameter mappings, in equation form, and log prior distribution of the parameters.
function [A,B,C,D,mean0,Cov0] = paramMap(params) a0 = params(1); a1 = params(2); b = params(3); d = params(4); A = @(x) max(0,a0+a1.*x); B = b; C = 1; D = d; mean0 = 0; Cov0 = 1; end function logprior = priorDistribution(theta) paramconstraints = theta(3:4) <= 0; % b and d are greater than 0 if(sum(paramconstraints)) logprior = -Inf; else logprior = 0; % Prior density is proportional to 1 for all values % in the parameter space. end end
Input Arguments
Observed response data, specified as a numeric matrix or a cell vector of numeric vectors.
If
PriorMdlis time invariant with respect to the observation equation,Yis a T-by-n matrix. Each row of the matrix corresponds to a period and each column corresponds to a particular observation in the model. T is the sample size and n is the number of observations per period. The last row ofYcontains the latest observations.If
PriorMdlis time varying with respect to the observation equation,Yis a T-by-1 cell vector.Y{t}contains an nt-dimensional vector of observations for period t, where t = 1, ..., T. For linear observation models, the corresponding dimensions of the coefficient matrices, outputs ofPriorMdl.ParamMap,C{t}, andD{t}must be consistent with the matrix inY{t}for all periods. For nonlinear observation models, the dimensions of the inputs and outputs associated with the observations must be consistent. Regardless of model type, the last cell ofYcontains the latest observations.
NaN elements indicate missing observations. For details on how
tune accommodates missing observations, see Algorithms.
Data Types: double | cell
Initial parameter values for the parameters Θ, specified as a
numParams-by-1 numeric vector. Elements of
params0 must correspond to the elements of the first input
arguments of PriorMdl.ParamMap and
PriorMdl.ParamDistribution.
Data Types: double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: tune(PriorMdl,Y,params0,Hessian="opg",Display="off") uses the
outer-product of gradients method to compute the Hessian matrix and suppresses the display
of the optimized values.
Number of particles for SMC, specified as a positive integer.
Example: NumParticles=1e4
Data Types: double
Since R2025a
SMC proposal (importance) distribution, specified as a value in this table.
| Value | SMC Sampler | Description |
|---|---|---|
"bootstrap" | Bootstrap forward filter [7] | SMC samples particles form the state transition distribution (observations do not inform the sampler). This option
is available only for models with observation noise (nonzero
This method has a relatively low computational cost, but it does not inform the routine of the current observations, which can increase the Monte Carlo sampling variance. |
"optimal" | Conditionally optimal proposal [4] | SMC samples particles from the one-step filtering distribution with incremental weights proportional to the likelihood (observations inform the sampler). The weights have minimum variance. This option is available for the following tractable models supplied in equation form:
|
"unscented" | Unscented transformation [11] | SMC proposal distribution is the one-step filtering distribution approximated by the unscented transformation (observations inform the sampler). This option is
available only for models in equation form with observation noise
(nonzero By default, this option
uses the filtered state mean as the representative point, around
which the algorithm generates sigma points. To specify all particles
as representative points for the unscented transformation, set
|
Example: NewSamples="unscented"
Data Types: char | string
Since R2025a
Flag to apply unscented transformation to all particles, specified as a value in this table.
| Value | Description |
|---|---|
false | The filtered state mean is the representative point, around which the algorithm generates sigma points. This
option is less computationally expensive than
|
true | All particles are the representative points. This option is more computationally expensive than the default, but it can yield a higher-quality proposal distribution. |
Example: MultiPassUnscented=false
Data Types: logical
Number of sample state paths to draw from the posterior smoothed state distribution, specified as a positive integer.
Example: NumPaths=1e4
Data Types: double
SMC resampling method, specified as a value in this table.
| Value | Description |
|---|---|
"multinomial" | At time t, the set of previously generated particles (parent set) follows a standard multinomial distribution, with probabilities proportional to their weights. An offspring set is resampled with replacement from the parent set [1]. |
"residual" | Residual sampling, a modified version of multinomial resampling that can produce an estimator with lower variance than the multinomial resampling method [9]. |
"systematic" | Systematic sampling, which produces an estimator with lower variance than the multinomial resampling method [3]. |
Resampling methods downsample insignificant particles to achieve a smaller estimator variance than if no resampling is performed and to avoid sampling from a degenerate proposal [5].
Example: Resample="residual"
Data Types: char | string
Effective sample size threshold, below which tune resamples particles, specified as a nonnegative scalar. For more details, see [5], Ch. 12.3.3.
Tip
To resample during every period, set
Cutoff=numparticles, wherenumparticlesis the value of theNumParticlesname-value argument.To avoid resampling, set
Cutoff=0.
Example: Cutoff=0.75*numparticles
Data Types: double
Flag for sorting particles before resampling, specified as a value in this table.
| Value | Description |
|---|---|
true | tune sorts the generated particles before resampling them. |
false | tune does not sort the generated particles. |
When SortPartiles=true, tune uses Hilbert sorting during the SMC routine to sort the particles. This action can reduce Monte Carlo variation, which is useful when you compare loglikelihoods resulting from evaluating several params arguments that are close to each other [3]. However, the sorting routine requires more computation resources, and can slow down computations, particularly in problems with a high-dimensional state variable.
Example: SortParticles=false
Data Types: logical
Parameter lower bounds when computing the Hessian matrix (see Hessian),
specified as a numParams-by-1 numeric vector.
Lower( specifies the lower bound of parameter j)theta(, the first input argument of j)PriorMdl.ParamMap and PriorMdl.ParamDistribution.
The default value [] specifies no lower bounds.
Note
Lower does not apply to posterior simulation. To apply parameter constraints on the posterior, code them in the log prior distribution function PriorMdl.ParamDistribution by setting the log prior of values outside the distribution support to -Inf.
Example: Lower=[0 -5 -1e7]
Data Types: double
Parameter lower bounds when computing the Hessian matrix (see Hessian),
specified as a numParams-by-1 numeric vector.
Upper( specifies the upper bound of parameter j)theta(, the first input argument of j)PriorMdl.ParamMap and PriorMdl.ParamDistribution.
The default value [] specifies no upper bounds.
Note
Upper does not apply to posterior simulation. To apply parameter constraints on the posterior, code them in the log prior distribution function PriorMdl.ParamDistribution by setting the log prior of values outside the distribution support to -Inf.
Example: Upper=[5 100 1e7]
Data Types: double
Optimization options for tuning the proposal distribution, specified as an optimoptions optimization controller. Options replaces default optimization options of the optimizer. For details on altering default values of the optimizer, see the optimization controller optimoptions, the constrained optimization function fmincon, or the unconstrained optimization function fminunc in Optimization Toolbox™.
For example, to change the constraint tolerance to 1e-6, set options = optimoptions(@fmincon,ConstraintTolerance=1e-6,Algorithm="sqp"). Then, pass Options by using Options=options.
By default, tune uses the default options of the optimizer.
Step size for computing the first derivatives that comprise the Hessian matrix (see
Hessian), specified as a positive scalar.
Example: StepSizeFirstDerivative=1e-4
Data Types: double
Step size for computing the second derivatives that comprise the Hessian matrix (see Hessian), specified as a positive scalar.
Example: StepSizeSecondDerivative=1e-3
Data Types: double
Hessian approximation method for the MH proposal distribution scale matrix, specified as a value in this table.
| Value | Description |
|---|---|
"difference" | Finite differencing |
"diagonal" | Diagonalized result of finite differencing |
"opg" | Outer product of gradients, ignoring the prior distribution |
Tip
The Hessian="difference" setting can be computationally intensive and inaccurate, and the resulting scale matrix can be nonnegative definite. Try one of the other options for better results.
Example: Hessian="opg"
Data Types: char | string
Control for displaying the proposal-tuning results, specified as a value in this table.
| Value | Description |
|---|---|
"off" | Suppress all output. |
"summary" | Display a summary of proposal-tuning results. |
"full" | Display proposal-tuning details. |
Example: Display="full"
Data Types: string | char
Output Arguments
Optimized parameter values for the MCMC sampler, returned as a
numParams-by-1 numeric vector.
params( contains the optimized value
of parameter j)theta(, where
j)theta is the first input argument of
PriorMdl.ParamMap and
PriorMdl.ParamDistribution.
When you call simulate, pass
params as the initial parameter values input
params0.
Proposal distribution covariance/scale matrix for the MCMC sampler, specified as a
numParams-by-numParams numeric matrix. Rows and
columns of Proposal correspond to elements in
params.
Proposal is the scale matrix up to a proportionality constant,
which is specified by the Proportion name-value argument of
estimate and
simulate.
The proposal distribution is multivariate normal or Student's t.
When you call simulate, pass
Proposal as the proposal distribution scale matrix input
Proposal.
Data Types: double
Tips
Unless you set
Cutoff=0,tuneresamples particles according to the specified resampling methodResample. Although resampling particles with high weights improves the results of the SMC, you should also allow the sampler traverse the proposal distribution to obtain novel, high-weight particles. To do this, experiment withCutoff.Avoid an arbitrary choice of the initial state distribution.
bnlssmfunctions generate the initial particles from the specified initial state distribution, which impacts the performance of the nonlinear filter. If the initial state specification is bad enough, importance weights concentrate on a small number of particles in the first SMC iteration, which might produce unreasonable filtering results. This vulnerability of the nonlinear model behavior contrasts with the stability of the Kalman filter for the linear model, in which the initial state distribution usually has little impact on the filter because the prior is washed out as it processes data.
Algorithms
The MCMC sampler requires a carefully specified proposal distribution.
tunetunes the sampler by performing numerical optimization to search for a posterior mode. A reasonable proposal for the multivariate normal or t distribution is the inverse of the negative Hessian matrix, whichtuneevaluates at the resulting posterior mode.tuneapproximates the data likelihood by sequential Monte Carlo (SMC), and it approximates the posterior gradient vector and Hessian matrix by the simulation smoother (simsmooth).tunetunes the sampler by numerical optimization.When
tunetunes the proposal distribution, the optimizer thattuneuses to search for a posterior mode before computing the Hessian matrix depends on your specifications.tuneaccommodates missing data by not updating filtered state estimates corresponding to missing observations. In other words, suppose there is a missing observation at period t. Then, the state forecast for period t based on the previous t – 1 observations and filtered state for period t are equivalent.
References
[1] Andrieu, Christophe, Arnaud Doucet, and Roman Holenstein. "Particle Markov Chain Monte Carlo Methods." Journal of the Royal Statistical Society Series B: Statistical Methodology 72 (June 2010): 269–342. https://doi.org/10.1111/j.1467-9868.2009.00736.x.
[2] Andrieu, Christophe, and Gareth O. Roberts. "The Pseudo-Marginal Approach for Efficient Monte Carlo Computations." Ann. Statist. 37 (April 2009): 697–725. https://dx.doi.org/10.1214/07-AOS574.
[3] Deligiannidis, George, Arnaud Doucet, and Michael Pitt. "The Correlated Pseudo-Marginal Method." Journal of the Royal Statistical Society, Series B: Statistical Methodology 80 (June 2018): 839–870. https://doi.org/10.1111/rssb.12280.
[4] Doucet, Arnaud, Simon Godsill, and Christophe Andrieu. "On Sequential Monte Carlo Sampling Methods for Bayesian Filtering." Statistics and Computing 10 (July 2000): 197–208. https://doi.org/10.1023/A:1008935410038.
[5] Durbin, J, and Siem Jan Koopman. Time Series Analysis by State Space Methods. 2nd ed. Oxford: Oxford University Press, 2012.
[6] Fernández-Villaverde, Jesús, and Juan F. Rubio-Ramírez. "Estimating Macroeconomic Models: A Likelihood Approach." Review of Economic Studies 70(October 2007): 1059–1087. https://doi.org/10.1111/j.1467-937X.2007.00437.x.
[7] Gordon, Neil J., David J. Salmond, and Adrian F. M. Smith. "Novel Approach to Nonlinear/Non-Gaussian Bayesian State Estimation." IEE Proceedings F Radar and Signal Processing 140 (April 1993): 107–113. https://doi.org/10.1049/ip-f-2.1993.0015.
[8] Hastings, Wilfred K. "Monte Carlo Sampling Methods Using Markov Chains and Their Applications." Biometrika 57 (April 1970): 97–109. https://doi.org/10.1093/biomet/57.1.97.
[9] Liu, Jun, and Rong Chen. "Sequential Monte Carlo Methods for Dynamic Systems." Journal of the American Statistical Association 93 (September 1998): 1032–1044. https://dx.doi.org/10.1080/01621459.1998.10473765.
[10] Metropolis, Nicholas, Rosenbluth, Arianna. W., Rosenbluth, Marshall. N., Teller, Augusta. H., and Teller, Edward. "Equation of State Calculations by Fast Computing Machines." The Journal of Chemical Physics 21 (June 1953): 1087–92. https://doi.org/10.1063/1.1699114.
[11] van der Merwe, Rudolph, Arnaud Doucet, Nando de Freitas, and Eric Wan. "The Unscented Particle Filter." Advances in Neural Information Processing Systems 13 (November 2000). https://dl.acm.org/doi/10.5555/3008751.3008833.
Version History
Introduced in R2024bWhile the SMC routine implements forward filtering, it resamples particles to align them to
the target distribution. The NewSamples name-value argument enables you
to choose how the SMC sampler chooses the particles. The supported algorithms are the
bootstrap forward filter ("bootstrap"), the conditionally optimal
proposal ("optimal"), and the unscented transformation
("unscented).
By default, the unscented transformation uses the filtered state mean as the representative
point, around which sigma points are generated. The MultiPassUnscented
name-value argument enables you to specify all particles as representative points for the
unscented transformation.
Before R2025a, the functions implement forward filtering using only the bootstrap filter.
See Also
Objects
Functions
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)