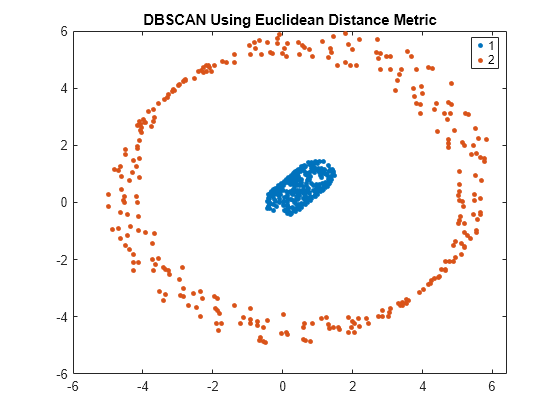
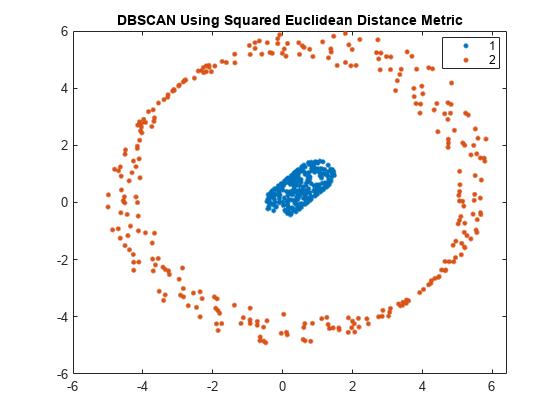
dbscan
Density-based spatial clustering of applications with noise (DBSCAN)
Syntax
Description
idx = dbscan(X,epsilon,minpts)X into clusters using the DBSCAN algorithm (see Algorithms).
dbscan clusters the observations (or points) based on a threshold
for a neighborhood search radius epsilon and a minimum number of
neighbors minpts required to identify a core point. The function
returns an n-by-1 vector (idx) containing cluster
indices of each observation.
idx = dbscan(D,epsilon,minpts,'Distance','precomputed')D between observations. D can be the output of
pdist or pdist2, or a more general dissimilarity vector or matrix conforming to the
output format of pdist or pdist2,
respectively.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
More About
Tips
For improved speed when iterating over many values of
epsilon, consider passing inDas the input todbscan. This approach prevents the function from having to compute the distances at every point of the iteration.If you use
pdist2to precomputeD, do not specify the'Smallest'or'Largest'name-value pair arguments ofpdist2to select or sort columns ofD. Selecting fewer than n distances results in an error, becausedbscanexpectsDto be a square matrix. Sorting the distances in each column ofDleads to a loss in the interpretation ofDand can give meaningless results when used in thedbscanfunction.For efficient memory usage, consider passing in
Das a logical matrix rather than a numeric matrix todbscanwhenDis large. By default, MATLAB® stores each value in a numeric matrix using 8 bytes (64 bits), and each value in a logical matrix using 1 byte (8 bits).To select a value for
minpts, consider a value greater than or equal to the number of dimensions of the input data plus one [1]. For example, for an n-by-p matrixX, set'minpts'equal to p+1 or greater.One possible strategy for selecting a value for
epsilonis to generate a k-distance graph forX. For each point inX, find the distance to the kth nearest point, and plot sorted points against this distance. Generally, the graph contains a knee. The distance that corresponds to the knee is typically a good choice forepsilon, because it is the region where points start tailing off into outlier (noise) territory [1].
Algorithms
DBSCAN is a density-based clustering algorithm that is designed to discover clusters and noise in data. The algorithm identifies three kinds of points: core points, border points, and noise points [1]. For specified values of
epsilonandminpts, thedbscanfunction implements the algorithm as follows:From the input data set
X, select the first unlabeled observation x1 as the current point, and initialize the first cluster label C to 1.Find the set of points within the epsilon neighborhood
epsilonof the current point. These points are the neighbors.If the number of neighbors is less than
minpts, then label the current point as a noise point (or an outlier). Go to step 4.Note
dbscancan reassign noise points to clusters if the noise points later satisfy the constraints set byepsilonandminptsfrom some other point inX. This process of reassigning points happens for border points of a cluster.Otherwise, label the current point as a core point belonging to cluster C.
Iterate over each neighbor (new current point) and repeat step 2 until no new neighbors are found that can be labeled as belonging to the current cluster C.
Select the next unlabeled point in
Xas the current point, and increase the cluster count by 1.Repeat steps 2–4 until all points in
Xare labeled.
If two clusters have varying densities and are close to each other, that is, the distance between two border points (one from each cluster) is less than
epsilon, thendbscancan merge the two clusters into one.Every valid cluster might not contain at least
minptsobservations. For example,dbscancan identify a border point belonging to two clusters that are close to each other. In such a situation, the algorithm assigns the border point to the first discovered cluster. As a result, the second cluster is still a valid cluster, but it can have fewer thanminptsobservations.
References
[1] Ester, M., H.-P. Kriegel, J. Sander, and X. Xiaowei. “A density-based algorithm for discovering clusters in large spatial databases with noise.” In Proceedings of the Second International Conference on Knowledge Discovery in Databases and Data Mining, 226-231. Portland, OR: AAAI Press, 1996.
Extended Capabilities
Version History
Introduced in R2019a