pdist2
两组观测值之间的两两距离
语法
说明
D = pdist2(X,Y,Distance,DistParameter)Distance 和 DistParameter 指定的度量返回该距离。仅当 Distance 是 'seuclidean'、'minkowski' 或 'mahalanobis' 时,您才能指定 DistParameter。
对于任何以前的参量,D = pdist2(___,Name,Value)
D = pdist2(X,Y,Distance,'Smallest',K)使用Distance指定的度量计算距离,并以升序返回X中观测值与Y中每个观测值的前K个最小两两距离。D = pdist2(X,Y,Distance,DistParameter,'Largest',K)使用Distance和DistParameter指定的度量计算距离,并以降序返回K个最大的两两距离。
示例
创建包含三个观测值和两个变量的两个矩阵。
rng('default') % For reproducibility X = rand(3,2); Y = rand(3,2);
计算欧几里德距离。输入参量 Distance 的默认值为 'euclidean'。在不使用名称-值对组参量的情况下计算欧几里德距离时,不需要指定 Distance。
D = pdist2(X,Y)
D = 3×3
0.5387 0.8018 0.1538
0.7100 0.5951 0.3422
0.8805 0.4242 1.2050
D(i,j) 对应于 X 中的观测值 i 与 Y 中的观测值 j 之间的两两距离。
创建包含三个观测值和两个变量的两个矩阵。
rng('default') % For reproducibility X = rand(3,2); Y = rand(3,2);
使用默认指数 2 计算闵可夫斯基距离。
D1 = pdist2(X,Y,'minkowski')D1 = 3×3
0.5387 0.8018 0.1538
0.7100 0.5951 0.3422
0.8805 0.4242 1.2050
用指数 1 计算闵可夫斯基距离,它等于城市街区距离。
D2 = pdist2(X,Y,'minkowski',1)D2 = 3×3
0.5877 1.0236 0.2000
0.9598 0.8337 0.3899
1.0189 0.4800 1.7036
D3 = pdist2(X,Y,'cityblock')D3 = 3×3
0.5877 1.0236 0.2000
0.9598 0.8337 0.3899
1.0189 0.4800 1.7036
创建两个包含五个观测值和两个变量的矩阵。
rng(0,"twister") % For reproducibility X = rand(5,2); Y = rand(5,2);
使用 pdist2 函数计算马氏距离。
D = pdist2(X,Y,"mahalanobis")D = 5×5
2.0012 0.9926 2.1767 1.9656 2.2036
2.3429 0.4318 1.6528 1.7564 1.7453
1.0330 2.5697 2.7833 1.3093 2.3936
3.2463 1.3676 0.1638 1.4094 0.3452
2.6608 1.6585 0.9895 0.6572 0.5121
计算 X 的均值与观测值 Y 之间的马氏距离。使用 X 的协方差作为距离度量参数。
D2 = pdist2(mean(X),Y,"mahalanobis",cov(X))D2 = 1×5
2.0090 0.9377 1.2824 0.7850 1.0966
使用 mahal 函数计算平方马氏距离。
SqMahalDist = mahal(Y,X)'
SqMahalDist = 1×5
4.0360 0.8792 1.6445 0.6162 1.2025
计算每个值的平方根。
MahalDist = SqMahalDist.^0.5
MahalDist = 1×5
2.0090 0.9377 1.2824 0.7850 1.0966
马氏距离值与 pdist2 函数返回的值相同。
创建包含三个观测值和两个变量的两个矩阵。
rng('default') % For reproducibility X = rand(3,2); Y = rand(3,2);
为 Y 中的每个观测值求出 X 中观测值的两个最小的两两欧几里德距离。
[D,I] = pdist2(X,Y,'euclidean','Smallest',2)
D = 2×3
0.5387 0.4242 0.1538
0.7100 0.5951 0.3422
I = 2×3
1 3 1
2 2 2
对于 Y 中的每个观测值,pdist2 通过计算距离值并将其与 X 中的所有观测值进行比较,求出两个最小的距离。然后,该函数按升序对 D 的每列中的距离进行排序。I 包含与 D 中的距离对应的 X 中的观测值的索引。
创建两个由点组成的大型矩阵,然后测量 pdist2 采用默认的 "euclidean" 距离度量时所用的时间。
rng default % For reproducibility N = 10000; X = randn(N,1000); Y = randn(N,1000); D = pdist2(X,Y); % Warm up function for more reliable timing information tic D = pdist2(X,Y); standard = toc
standard = 12.2637
接下来,使用 "fasteuclidean" 距离度量来测量 pdist2 所用的时间。指定缓存大小为 100。
D = pdist2(X,Y,"fasteuclidean",CacheSize=100); % Warm up function tic D2 = pdist2(X,Y,"fasteuclidean",CacheSize=100); accelerated = toc
accelerated = 2.2260
计算加速后的计算比标准计算快多少倍。
standard/accelerated
ans = 5.5094
在此示例中,加速版本的速度提高了一倍多。
定义一个忽略 NaN 值坐标的自定义距离函数,并使用该自定义距离函数计算两两距离。
创建包含三个观测值和三个变量的两个矩阵。
rng('default') % For reproducibility X = rand(3,3) Y = [X(:,1:2) rand(3,1)]
X =
0.8147 0.9134 0.2785
0.9058 0.6324 0.5469
0.1270 0.0975 0.9575
Y =
0.8147 0.9134 0.9649
0.9058 0.6324 0.1576
0.1270 0.0975 0.9706
X 和 Y 的前两列是相同的。假设缺少 X(1,1)。
X(1,1) = NaN
X =
NaN 0.9134 0.2785
0.9058 0.6324 0.5469
0.1270 0.0975 0.9575
计算汉明距离。
D1 = pdist2(X,Y,'hamming')
D1 =
NaN NaN NaN
1.0000 0.3333 1.0000
1.0000 1.0000 0.3333
如果 X 中的观测值 i 或 Y 中的观测值 j 包含 NaN 值,则对于 i 和 j 之间的两两距离,函数 pdist2 返回 NaN。因此,D1(1,1)、D1(1,2) 和 D1(1,3) 为 NaN 值。
定义一个自定义距离函数 nanhamdist,该函数忽略 NaN 值的坐标并计算汉明距离。当处理大量观测值时,您可以通过遍历数据的坐标来更快地计算距离。
function D2 = nanhamdist(XI,XJ) %NANHAMDIST Hamming distance ignoring coordinates with NaNs [m,p] = size(XJ); nesum = zeros(m,1); pstar = zeros(m,1); for q = 1:p notnan = ~(isnan(XI(q)) | isnan(XJ(:,q))); nesum = nesum + ((XI(q) ~= XJ(:,q)) & notnan); pstar = pstar + notnan; end D2 = nesum./pstar;
将函数句柄作为输入参量传递给 pdist2,以使用 nanhamdist 计算该距离。
D2 = pdist2(X,Y,@nanhamdist)
D2 =
0.5000 1.0000 1.0000
1.0000 0.3333 1.0000
1.0000 1.0000 0.3333
kmeans 执行 k 均值聚类以将数据划分为 k 个簇。当您有要进行聚类的新数据集时,可以使用 kmeans 创建包含现有数据和新数据的新簇。kmeans 函数支持 C/C++ 代码生成,因此您可以生成接受训练数据并返回聚类结果的代码,然后将代码部署到设备上。在此工作流中,您必须传递训练数据,训练数据有可能相当大。为了节省设备上的内存,您可以分别使用 kmeans 和 pdist2 来分离训练和预测。
使用 kmeans 在 MATLAB® 中创建簇,并在生成的代码中使用 pdist2 将新数据分配给现有簇。对于代码生成,定义接受簇质心位置和新数据集的入口函数,并返回最近邻簇的索引。然后,为入口函数生成代码。
生成 C/C++ 代码需要 MATLAB® Coder™。
执行 k 均值聚类
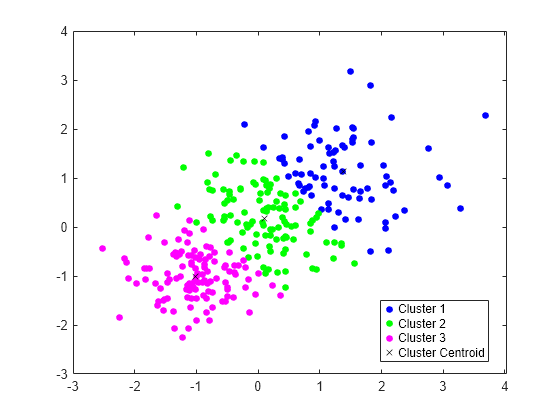
使用三种分布生成训练数据集。
rng('default') % For reproducibility X = [randn(100,2)*0.75+ones(100,2); randn(100,2)*0.5-ones(100,2); randn(100,2)*0.75];
使用 kmeans 将训练数据分成三个簇。
[idx,C] = kmeans(X,3);
绘制簇和簇质心。
figure gscatter(X(:,1),X(:,2),idx,'bgm') hold on plot(C(:,1),C(:,2),'kx') legend('Cluster 1','Cluster 2','Cluster 3','Cluster Centroid')
将新数据分配给现有簇
生成测试数据集。
Xtest = [randn(10,2)*0.75+ones(10,2);
randn(10,2)*0.5-ones(10,2);
randn(10,2)*0.75];使用现有簇对测试数据集进行分类。使用 pdist2 找到距离每个测试数据点最近的质心。
[~,idx_test] = pdist2(C,Xtest,'euclidean','Smallest',1);
使用 idx_test 和 gscatter 绘制测试数据并对测试数据加标签。
gscatter(Xtest(:,1),Xtest(:,2),idx_test,'bgm','ooo') legend('Cluster 1','Cluster 2','Cluster 3','Cluster Centroid', ... 'Data classified to Cluster 1','Data classified to Cluster 2', ... 'Data classified to Cluster 3')

生成代码
生成将新数据分配给现有簇的 C 代码。请注意,生成 C/C++ 代码需要 MATLAB® Coder™。
定义名为 findNearestCentroid 的入口函数,该函数接受质心位置和新数据,然后使用 pdist2 找到最近的簇。
在入口函数的函数签名后面添加 %#codegen 编译器指令(即 pragma),以指示您要为此 MATLAB 算法生成代码。添加此指令指示 MATLAB 代码分析器帮助您诊断和修复在代码生成过程中可能导致错误的违规。
type findNearestCentroid % Display contents of findNearestCentroid.m
function idx = findNearestCentroid(C,X) %#codegen [~,idx] = pdist2(C,X,'euclidean','Smallest',1); % Find the nearest centroid
注意:如果您点击位于此页右上角的按钮,并在 MATLAB® 中打开此示例,则 MATLAB® 将打开示例文件夹。该文件夹包括入口函数文件。
使用 codegen (MATLAB Coder) 生成代码。由于 C 和 C++ 是静态类型语言,因此必须在编译时确定入口函数中所有变量的属性。要指定 findNearestCentroid 的输入的数据类型和数组大小,请使用 -args 选项传递表示具有特定数据类型和数组大小的值集的 MATLAB 表达式。有关详细信息,请参阅 Specify Variable-Size Arguments for Code Generation。
codegen findNearestCentroid -args {C,Xtest}
Code generation successful.
codegen 生成 MEX 函数 findNearestCentroid_mex,扩展名因平台而异。
验证生成的代码。
myIndx = findNearestCentroid(C,Xtest); myIndex_mex = findNearestCentroid_mex(C,Xtest); verifyMEX = isequal(idx_test,myIndx,myIndex_mex)
verifyMEX = logical
1
isequal 返回逻辑值 1 (true),这意味着所有输入都相等。这一比较结果确认 pdist2 函数、findNearestCentroid 函数和 MEX 函数均返回相同的索引。
您还可以使用 GPU Coder™ 生成优化的 CUDA® 代码。
cfg = coder.gpuConfig('mex'); codegen -config cfg findNearestCentroid -args {C,Xtest}
有关代码生成的详细信息,请参阅General Code Generation Workflow。有关 GPU Coder 的详细信息,请参阅Get Started with GPU Coder (GPU Coder) 和Supported Functions (GPU Coder)。
输入参数
输入数据,指定为数值矩阵。X 是一个 mx×n 矩阵,而 Y 是一个 my×n 矩阵。行对应于单个观测值,列对应单个变量。
数据类型: single | double
距离度量,指定为字符向量、字符串标量或函数句柄,如下表中所述。
| 值 | 描述 |
|---|---|
'euclidean' | 欧几里德距离(默认值) |
'squaredeuclidean' | 平方欧几里德距离。(此选项仅用于提高效率。它不满足三角不等式。) |
'seuclidean' | 标准化的欧几里德距离。每个观测值间坐标差都通过除以标准差 |
'fasteuclidean' | 当预测变量的数目至少为 10 时,使用替代算法计算的欧几里德距离,该算法可以节省时间。在某些情况下,这种更快的算法会降低准确度。以 'fast' 开头的算法不支持稀疏数据。有关详细信息,请参阅 算法。 |
'fastsquaredeuclidean' | 当预测变量的数目至少为 10 时,使用替代算法计算的平方欧几里德距离,该算法可以节省时间。在某些情况下,这种更快的算法会降低准确度。以 'fast' 开头的算法不支持稀疏数据。有关详细信息,请参阅 算法。 |
'fastseuclidean' | 当预测变量的数目至少为 10 时,使用替代算法计算的标准化的欧几里德距离,该算法可以节省时间。在某些情况下,这种更快的算法会降低准确度。以 'fast' 开头的算法不支持稀疏数据。有关详细信息,请参阅 算法。 |
'mahalanobis' | 马氏距离,使用 |
'cityblock' | 城市街区距离 |
'minkowski' | 闵可夫斯基距离。默认指数是 2。使用 |
'chebychev' | 切比雪夫距离(最大坐标差) |
'cosine' | 1 减去点之间夹角的余弦值(视为向量) |
'correlation' | 1 减去点之间的样本相关性(视为值序列) |
'hamming' | 汉明距离,即相异坐标所占的百分比 |
'jaccard' | 1 减去杰卡德系数,即非零相异坐标所占的百分比 |
'spearman' | 1 减去样本观测值(视为值序列)之间的斯皮尔曼秩相关 |
@ | 自定义距离函数句柄。距离函数的形式如下 function D2 = distfun(ZI,ZJ) % calculation of distance ...
对于非稀疏数据,使用内置距离度量计算距离通常比使用函数句柄更快。 |
有关定义,请参阅距离度量。
当您使用 'seuclidean'、'minkowski' 或 'mahalanobis' 时,您可以指定额外的输入参量 DistParameter 来控制这些度量。您也可以像使用其他度量一样来使用这些度量,但这种情况下使用的是 DistParameter 的默认值。
示例: 'minkowski'
数据类型: char | string | function_handle
距离度量参数值,指定为正标量、数值向量或数值矩阵。仅当您将 Distance 指定为 'seuclidean'、'minkowski' 或 'mahalanobis' 时,此参量才有效。
如果
Distance是'seuclidean',DistParameter是对应于每个维度的缩放因子的向量,指定为正向量。默认值为std(X,'omitnan')。如果
Distance是'minkowski',DistParameter是闵可夫斯基距离的指数,指定为正标量。默认值为 2。如果
Distance是'mahalanobis',DistParameter是协方差矩阵,指定为数值矩阵。默认值为cov(X,'omitrows')。DistParameter必须是对称正定矩阵。
示例: 'minkowski',3
数据类型: single | double
名称-值参数
将可选参量对组指定为 Name1=Value1,...,NameN=ValueN,其中 Name 是参量名称,Value 是对应的值。名称-值参量必须出现在其他参量之后,但对各个参量对组的顺序没有要求。
在 R2021a 之前,使用逗号分隔每个名称和值,并用引号将 Name 引起来。
示例: 或者使用 'Smallest',K,或者使用 'Largest',K。但您不能同时使用 'Smallest' 和 'Largest'。
格拉姆矩阵的大小,以 MB 为单位,指定为正标量或 'maximal'。仅当 Distance 参量以 fast 开头时,pdist2 函数才能使用 CacheSize。
如果是 'maximal',pdist2 尝试为大小为 MX×MY 的整个中间矩阵分配足够的内存,其中 MX 是输入数据 X 的行数,MY 是输入数据 Y 的行数。高速缓存的大小不必大到足以容纳整个中间矩阵,但必须至少大到足以容纳一个 MX×1 向量。否则,pdist2 使用常规算法来计算欧几里德距离。
如果距离参量以 fast 开头,并且 CacheSize 太大或为 'maximal',则 pdist2 可能会尝试分配超出可用内存的格拉姆矩阵。在这种情况下,MATLAB® 会引发错误。
示例: CacheSize='maximal'
数据类型: double | char | string
要求出的最小距离的数目,指定为由 'Smallest' 和一个正整数组成的以逗号分隔的对组。如果指定 'Smallest',则 pdist2 以升序对 D 的每列中的距离进行排序。您只能使用参量 Smallest 和 Largest 之一。
示例: 'Smallest',3
数据类型: single | double
要求出的最大距离的数目,指定为由 'Largest' 和一个正整数组成的以逗号分隔的对组。如果指定 'Largest',则 pdist2 以降序对 D 的每列中的距离进行排序。您只能使用参量 Smallest 和 Largest 之一。
示例: 'Largest',3
数据类型: single | double
输出参量
两两距离,以数值矩阵形式返回。
如果未指定 'Smallest' 或 'Largest',则 D 是 mx×my 矩阵,其中 mx 和 my 分别是 X 和 Y 中的观测值数目。D(i,j) 是 X 中的观测值 i 与 Y 中的观测值 j 之间的距离。如果 X 中的观测值 i 或 Y 中的观测值 j 包含 NaN,则内置距离函数返回的 D(i,j) 为 NaN。
如果将 'Smallest' 或 'Largest' 指定为 K,则 D 是 K×my 矩阵。D 包含 Y 中每个观测值到 X 中观测值的 K 个最小或 K 个最大两两距离。对于 Y 中的每个观测值,pdist2 通过计算并比较其与 X 中所有观测值的距离值,求出 K 个最小或最大距离。如果 K 大于 mx,则 pdist2 返回一个 mx×my 矩阵。
详细信息
距离度量是定义两个观测值之间距离的函数。pdist2 支持各种距离度量:欧几里德距离、标准化的欧几里德距离、马氏距离、城市街区距离、闵可夫斯基距离、切比雪夫距离、余弦距离、相关性距离、汉明距离、杰卡德距离和斯皮尔曼距离。
给定一个 mx×n 数据矩阵 X(它被视为 mx 个 (1×n) 的行向量 x1、x2、...、xmx)和一个 my×n 数据矩阵 Y(它被视为 my 个 (1×n) 的行向量 y1、y2、...、ymy),则向量 xs 和 yt 之间的各种距离定义如下:
欧几里德距离
欧几里德距离是闵可夫斯基距离的特例,其中 p = 2。
标准化的欧几里德距离
其中 V 是 n×n 对角矩阵,它的第 j 个对角线元素是 (S(j))2,其中 S 是对应于每个维度的缩放因子的向量。
快速欧几里德距离与欧几里德距离相同,只是当预测变量的数目至少为 10 时会使用替代算法进行计算,该算法可以节省时间。在某些情况下,这种更快的算法会降低准确度。不支持稀疏数据。请参阅 快速欧几里德距离算法。
通过将
Distance参数设置为'fasteuclidean'可指定快速欧几里德距离。快速标准化的欧几里德距离与标准化的欧几里德距离相同,只是当预测变量的数目至少为 10 时会使用替代算法进行计算,该算法可以节省时间。在某些情况下,这种更快的算法会降低准确度。不支持稀疏数据。请参阅 快速欧几里德距离算法。
通过将
Distance参数设置为'fastseuclidean'可指定快速标准化的欧几里德距离。马氏距离
其中 C 是协方差矩阵。
城市街区距离
城市街区距离是闵可夫斯基距离的特例,其中 p = 1。
闵可夫斯基距离
对于特例 p = 1,闵可夫斯基距离即城市街区距离。对于特例 p = 2,闵可夫斯基距离即欧几里德距离。对于特例 p = ∞,闵可夫斯基距离即切比雪夫距离。
切比雪夫距离
切比雪夫距离是闵可夫斯基距离的特例,其中 p = ∞。
余弦距离
相关性距离
其中
且
汉明距离
汉明距离是相异坐标所占的百分比。
杰卡德距离是 1 减去杰卡德系数,即非零相异坐标所占的百分比:
斯皮尔曼距离是 1 减去样本观测值之间的斯皮尔曼秩相关(视为值序列):
其中
算法
以 fast 开头的 Distance 参量(如 "fasteuclidean" 和 "fastseuclidean")的值在计算欧几里德距离时使用的算法会使用额外的内存来节省计算时间。此算法在 Albanie 的文献 [1] 中和其他位置称为“欧几里德距离矩阵技巧”。内部测试表明,当预测变量的数目至少为 10 时,该算法可以节省时间。以 fast 开头的算法不支持稀疏数据。
为了求得所有点 xi 和 xj 之间距离的矩阵 D(其中每个 xi 具有 n 个变量),该算法使用以下方程中的最后一行来计算距离:
方程最后一行中的矩阵 称为格拉姆矩阵。当您计算并使用格拉姆矩阵而不是通过平方与求和来计算平方距离时,计算平方距离集的速度更快,但在数值上稳定性稍差。有关详细信息,请参阅 Albanie [1]。
为了存储格拉姆矩阵,软件使用默认大小为 1e3 MB 的缓存。您可以使用 CacheSize 名称-值参量设置缓存大小。如果 CacheSize 的值太大或为 "maximal",则软件可能会尝试分配超出可用内存容量的格拉姆矩阵。在这种情况下,软件会报错。
参考
[1] Albanie, Samuel. Euclidean Distance Matrix Trick. June, 2019. Available at https://samuelalbanie.com/files/Euclidean_distance_trick.pdf.
扩展功能
用法说明和限制:
距离输入参量值 (
Distance) 必须为编译时常量。例如,要使用闵可夫斯基距离,请将coder.Constant('Minkowski')包含在codegen的-args值中。距离输入参量值 (
Distance) 不能为自定义距离函数。pdist2不支持快速欧几里德距离计算的代码生成,即名称以fast开头的那些距离度量(例如'fasteuclidean')。名称-值参量中的名称必须为编译时常量。例如,要在生成的代码中使用
'Smallest'名称-值对组参量,请在codegen(MATLAB Coder) 的-args值中包括{coder.Constant('Smallest'),0}。由于数值精度的原因,生成的代码中相等距离的排序顺序可能不同于 MATLAB 中的顺序。
pdist2的生成代码使用parfor(MATLAB Coder) 在生成的代码中创建可在受支持的共享内存多核平台上并行运行的循环。如果您的编译器不支持 Open Multiprocessing (OpenMP) 应用程序接口,或您禁用 OpenMP 库,则 MATLAB Coder™ 会将parfor循环视为for循环。要查找支持的编译器,请参阅支持的编译器。要禁用 OpenMP 库,请将配置对象的EnableOpenMP属性设置为false。有关详细信息,请参阅coder.CodeConfig(MATLAB Coder)。pdist2在生成的独立 C/C++ 代码中返回整数类型 (int32) 索引。因此,当您使用单精度输入时,该函数允许严格的单精度支持。对于 MEX 代码生成,该函数仍返回双精度索引以匹配 MATLAB 行为。
有关代码生成的详细信息,请参阅 Introduction to Code Generation 和 General Code Generation Workflow。
用法说明和限制:
优化的 CUDA 代码支持的距离输入参量值 (
Distance) 包括'euclidean'、'squaredeuclidean'、'seuclidean'、'cityblock'、'minkowski'、'chebychev'、'cosine'、'correlation'、'hamming'和'jaccard'。Distance不能为自定义距离函数。Distance必须为编译时常量。名称-值对组参量中的名称必须为编译时常量。
由于数值精度的原因,生成的代码中相等距离的排序顺序可能不同于 MATLAB 中的顺序。
用法说明和限制:
不能将
Distance输入参量指定为"fasteuclidean"、"fastsquaredeuclidean"、"fastseuclidean"或自定义距离函数。
有关详细信息,请参阅在 GPU 上运行 MATLAB 函数 (Parallel Computing Toolbox)。
版本历史记录
在 R2010a 中推出另请参阅
pdist | createns | knnsearch | ExhaustiveSearcher | KDTreeSearcher
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)