kmedoids
k-medoids clustering
Syntax
Description
idx = kmedoids(X,k)X into k clusters, and returns an
n-by-1 vector idx containing cluster
indices of each observation. Rows of X correspond to points and
columns correspond to variables. By default, kmedoids uses
squared Euclidean distance metric and the k-means++
algorithm for choosing initial cluster medoid positions.
idx = kmedoids(X,k,Name,Value)Name,Value pair
arguments.
Examples
Randomly generate data.
rng('default'); % For reproducibility X = [randn(100,2)*0.75+ones(100,2); randn(100,2)*0.55-ones(100,2)]; figure; plot(X(:,1),X(:,2),'.'); title('Randomly Generated Data');

Group data into two clusters using kmedoids. Use the cityblock distance metric.
opts = statset('Display','iter'); [idx,C,sumd,d,midx,info] = kmedoids(X,2,'Distance','cityblock','Options',opts);
rep iter sum
1 1 209.856
1 2 209.856
Best total sum of distances = 209.856
info is a struct that contains information about how the algorithm was executed. For example, bestReplicate field indicates the replicate that was used to produce the final solution. In this example, the replicate number 1 was used since the default number of replicates is 1 for the default algorithm, which is pam in this case.
info
info = struct with fields:
algorithm: 'pam'
start: 'plus'
distance: 'cityblock'
iterations: 2
bestReplicate: 1
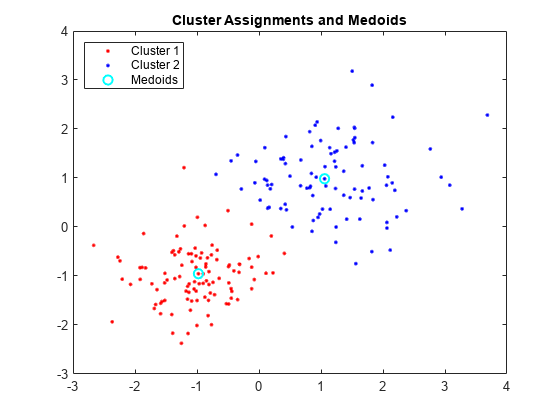
Plot the clusters and the cluster medoids.
figure; plot(X(idx==1,1),X(idx==1,2),'r.','MarkerSize',7) hold on plot(X(idx==2,1),X(idx==2,2),'b.','MarkerSize',7) plot(C(:,1),C(:,2),'co',... 'MarkerSize',7,'LineWidth',1.5) legend('Cluster 1','Cluster 2','Medoids',... 'Location','NW'); title('Cluster Assignments and Medoids'); hold off
This example uses "Mushroom" data set [3][4][5]
[6][7] from the UCI machine learning archive [7], described in https://archive.ics.uci.edu/dataset/73/mushroom. The data
set includes 22 predictors for 8,124 observations of various mushrooms. The
predictors are categorical data types. For example, cap shape is categorized
with features of 'b' for bell-shaped cap and
'c' for conical. Mushroom color is also categorized
with features of 'n' for brown, and
'p' for pink. The data set also includes a
classification for each mushroom of either edible or poisonous.
Since the features of the mushroom data set are categorical, it is not possible to define the mean of several data points, and therefore the widely-used k-means clustering algorithm cannot be meaningfully applied to this data set. k-medoids is a related algorithm that partitions data into k distinct clusters, by finding medoids that minimize the sum of dissimilarities between points in the data and their nearest medoid.
The medoid of a set is a member of that set whose average dissimilarity with the other members of the set is the smallest. Similarity can be defined for many types of data that do not allow a mean to be calculated, allowing k-medoids to be used for a broader range of problems than k-means.
Using k-medoids, this example clusters the mushrooms into two groups, based on the predictors provided. It then explores the relationship between those clusters and the classifications of the mushrooms as either edible or poisonous.
This example assumes that you have downloaded the "Mushroom" data set
[3][4][5]
[6][7] from the UCI database (https://archive.ics.uci.edu/dataset/73/mushroom) and
saved the text files agaricus-lepiota.data and agaricus-lepiota.names in
your current directory. There is no column header line in the data, so
readtable uses the default variable names.
clear all data = readtable('agaricus-lepiota.data','ReadVariableNames',false);
Display the first 5 mushrooms with their first few features.
data(1:5,1:10)
ans =
Var1 Var2 Var3 Var4 Var5 Var6 Var7 Var8 Var9 Var10
____ ____ ____ ____ ____ ____ ____ ____ ____ _____
'p' 'x' 's' 'n' 't' 'p' 'f' 'c' 'n' 'k'
'e' 'x' 's' 'y' 't' 'a' 'f' 'c' 'b' 'k'
'e' 'b' 's' 'w' 't' 'l' 'f' 'c' 'b' 'n'
'p' 'x' 'y' 'w' 't' 'p' 'f' 'c' 'n' 'n'
'e' 'x' 's' 'g' 'f' 'n' 'f' 'w' 'b' 'k'Extract the first column, labeled data for edible and poisonous groups. Then delete the column.
labels = data(:,1);
labels = categorical(labels{:,:});
data(:,1) = [];Store the names of predictors (features), which are described in agaricus-lepiota.names.
VarNames = {'cap_shape' 'cap_surface' 'cap_color' 'bruises' 'odor' ...
'gill_attachment' 'gill_spacing' 'gill_size' 'gill_color' ...
'stalk_shape' 'stalk_root' 'stalk_surface_above_ring' ...
'stalk_surface_below_ring' 'stalk_color_above_ring' ...
'stalk_color_below_ring' 'veil_type' 'veil_color' 'ring_number' ....
'ring_type' 'spore_print_color' 'population' 'habitat'};Set the variable names.
data.Properties.VariableNames = VarNames;
There are a total of 2480 missing values denoted as '?'.
sum(char(data{:,:}) == '?')ans =
2480Based on the inspection of the data set and its description,
the missing values belong only to the 11th variable (stalk_root).
Remove the column from the table.
data(:,11) = [];
kmedoids only accepts numeric data.
You need to cast the categories you have into numeric type. The distance
function you will use to define the dissimilarity of the data will
be based on the double representation of the categorical data.
cats = categorical(data{:,:});
data = double(cats);kmedoids can use any distance metric
supported by pdist2 to cluster. For this example you will cluster
the data using the Hamming distance because this is an appropriate
distance metric for categorical data as illustrated below. The Hamming
distance between two vectors is the percentage of the vector components
that differ. For instance, consider these two vectors.
v1 = [1 0 2 1];
v2 = [1 1 2 1];
They are equal in the 1st, 3rd and 4th coordinate. Since 1 of the 4 coordinates differ, the Hamming distance between these two vectors is .25.
You can use the function pdist2 to measure
the Hamming distance between the first and second row of data, the
numerical representation of the categorical mushroom data. The value
.2857 means that 6 of the 21 features of the mushroom differ.
pdist2(data(1,:),data(2,:),'hamming')ans =
0.2857In this example, you’re clustering the mushroom
data into two clusters based on features to see if the clustering
corresponds to edibility. The kmedoids function
is guaranteed to converge to a local minima of the clustering criterion;
however, this may not be a global minimum for the problem. It is a
good idea to cluster the problem a few times using the 'replicates' parameter.
When 'replicates' is set to a value, n,
greater than 1, the k-medoids algorithm is run n times,
and the best result is returned.
To run kmedoids to cluster data into 2
clusters, based on the Hamming distance and to return the best result
of 3 replicates, you run the following.
rng('default'); % For reproducibility [IDX, C, SUMD, D, MIDX, INFO] = kmedoids(data,2,'distance','hamming','replicates',3);
Let's assume that mushrooms in the predicted group 1 are poisonous and group 2 are all edible. To determine the performance of clustering results, calculate how many mushrooms in group 1 are indeed poisonous and group 2 are edible based on the known labels. In other words, calculate the number of false positives, false negatives, as well as true positives and true negatives.
Construct a confusion matrix (or matching matrix), where the
diagonal elements represent the number of true positives and true
negatives, respectively. The off-diagonal elements represent false
negatives and false positives, respectively. For convenience, use
the confusionmat function, which calculates a
confusion matrix given known labels and predicted labels. Get the
predicted label information from the IDX variable. IDX contains
values of 1 and 2 for each data point, representing poisonous and
edible groups, respectively.
predLabels = labels; % Initialize a vector for predicted labels. predLabels(IDX==1) = categorical({'p'}); % Assign group 1 to be poisonous. predLabels(IDX==2) = categorical({'e'}); % Assign group 2 to be edible. confMatrix = confusionmat(labels,predLabels)
confMatrix =
4176 32
816 3100Out of 4208 edible mushrooms, 4176 were correctly predicted to be in group 2 (edible group), and 32 were incorrectly predicted to be in group 1 (poisonous group). Similarly, out of 3916 poisonous mushrooms, 3100 were correctly predicted to be in group 1 (poisonous group), and 816 were incorrectly predicted to be in group 2 (edible group).
Given this confusion matrix, calculate the accuracy, which is the proportion of true results (both true positives and true negatives) against the overall data, and precision, which is the proportion of the true positives against all the positive results (true positives and false positives).
accuracy = (confMatrix(1,1)+confMatrix(2,2))/(sum(sum(confMatrix)))
accuracy =
0.8956precision = confMatrix(1,1) / (confMatrix(1,1)+confMatrix(2,1))
precision =
0.8365The results indicated that applying the k-medoids algorithm to the categorical features of mushrooms resulted in clusters that were associated with edibility.
Input Arguments
Data, specified as a numeric matrix. The rows of X correspond
to observations, and the columns correspond to variables.
Number of medoids in the data, specified as a positive integer.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: 'Distance','euclidean','Replicates',3,'Options',statset('UseParallel',1) specifies
Euclidean distance, three replicate medoids at different starting
values, and to use parallel computing.
Algorithm to find medoids, specified as the comma-separated
pair consisting of 'Algorithm' and 'pam', 'small', 'clara',
or 'large'. The default algorithm depends on the
number of rows of X.
If the number of rows of
Xis less than 3000,'pam'is the default algorithm.If the number of rows is between 3000 and 10000,
'small'is the default algorithm.For all other cases,
'large'is the default algorithm.
You can override the default choice by explicitly stating the algorithm. This table summarizes the available algorithms.
| Algorithm | Description |
|---|---|
'pam' |
Partitioning Around Medoids (PAM) is the classical
algorithm for solving the
k-medoids problem described in
[1]. After applying the initialization function to
select initial medoid positions, the program
performs the swap-step of the PAM algorithm, that
is, it searches over all possible swaps between
medoids and non-medoids to see if the sum of medoid
to cluster member distances goes down. You can
specify which initialization function to use via the
The algorithm proceeds as follows.
The algorithm iterates the build- and swap-steps until the medoids do not change, or other termination criteria are met. The algorithm can produce better solutions than the other algorithms in some situations, but it can be prohibitively long running. |
'small' |
Use an algorithm similar to the k-means algorithm
to find The algorithm proceeds as follows.
The algorithm repeats these steps until no further
updates occur or other termination criteria are met.
The algorithm has an optional PAM-like online update
phase (specified by the
|
'clara' | Clustering Large Applications (CLARA) [1] repeatedly performs the PAM algorithm on random subsets of the data. It aims to overcome scaling challenges posed by the PAM algorithm through sampling. The algorithm proceeds as follows.
The algorithm repeats these steps until the medoids do not change, or other termination criteria are met. For the best performance, it is
recommended that you perform multiple replicates. By
default, the program performs five replicates. Each
replicate samples |
'large' |
This is similar to the |
Example: 'Algorithm','pam'
A flag to perform PAM-like online update phase, specified as
a comma-separated pair consisting of 'OnlinePhase' and 'on' or 'off'.
If it is on, then kmedoids performs
a PAM-like update to the medoids after the Lloyd iterations in the small and large algorithms.
During this online update phase, the algorithm chooses a small subset
of data points in each cluster that are the furthest from and nearest
to medoid. For each chosen point, it reassigns the clustering of the
entire data set and check if this creates a smaller sum of distances
than the best known.
In other words, the swap considerations are limited to the points near the medoids and far from the medoids. The near points are considered in order to refine the clustering. The far points are considered in order to escape local minima. Turning on this feature tends to improve the quality of solutions generated by both algorithms. Total run time tends to increase as well, but the increase typically is less than one iteration of PAM.
Example: OnlinePhase,'off'
Distance metric, specified as the name of a distance metric described
in the following table, or a function handle.
kmedoids minimizes the sum of medoid to cluster
member distances.
| Value | Description |
|---|---|
'sqEuclidean' | Squared Euclidean distance (default) |
'euclidean' | Euclidean distance |
'seuclidean' | Standardized Euclidean distance. Each
coordinate difference between observations is scaled
by dividing by the corresponding element of the
standard deviation, |
'cityblock' | City block distance |
'minkowski' | Minkowski distance. The exponent is 2. |
'chebychev' | Chebychev distance (maximum coordinate difference) |
'mahalanobis' | Mahalanobis distance using the sample
covariance of |
'cosine' | One minus the cosine of the included angle between points (treated as vectors) |
'correlation' | One minus the sample correlation between points (treated as sequences of values) |
'spearman' | One minus the sample Spearman's rank correlation between observations (treated as sequences of values) |
'hamming' | Hamming distance, which is the percentage of coordinates that differ |
'jaccard' | One minus the Jaccard coefficient, which is the percentage of nonzero coordinates that differ |
@ | Custom distance function handle. A distance function has the form function D2 = distfun(ZI,ZJ) % calculation of distance ...
If your data is not sparse, you can generally compute distance more quickly by using a built-in distance instead of a function handle. |
For the definition of each distance metric, see Distance Metrics.
Example: 'Distance','hamming'
Options to control the iterative algorithm to minimize fitting criteria, specified as the
comma-separated pair consisting of 'Options' and a
structure array returned by statset. Supported fields
of the structure array specify options for controlling the iterative
algorithm. This table summarizes the supported fields.
| Field | Description |
|---|---|
Display | Level of display output. Choices are
'off' (default) and
'iter'. |
MaxIter | Maximum number of iterations allowed. The default
is 100. |
UseParallel | If true, compute in parallel.
If Parallel Computing Toolbox™ is not available, then computation
occurs in serial mode. The default is
false, meaning serial
computation. |
UseSubstreams | Set to true to compute in a
reproducible fashion. The default is
false. To compute reproducibly,
you must also set Streams to a
type allowing substreams:
'mlfg6331_64' or
'mrg32k3a'. |
Streams | A RandStream object or cell
array of such objects. |
For details about these options and parallel computing in Statistics and Machine Learning Toolbox™, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox) and Reproducibility in Parallel Statistical Computations.
Example: 'Options',statset('Display','off')
Number of times to repeat clustering using new initial cluster
medoid positions, specified as a positive integer. The default value
depends on the choice of algorithm. For pam and small,
the default is 1. For clara, the default is 5.
For large, the default is 3.
Example: 'Replicates',4
Number of samples to take from the data when executing the
clara algorithm, specified as a positive integer.
The default number of samples is calculated as
40+2*k.
Example: 'NumSamples',160
Percent of the data set to examine using the large algorithm,
specified as a positive number.
The program examines percentneighbors*size(X,1) number
of neighbors for the medoids. If there is no improvement in the within-cluster
sum of distances, then the algorithm terminates.
The value of this parameter between 0 and 1, where a value closer to 1 tends to give higher quality solutions, but the algorithm takes longer to run, and a value closer to 0 tends to give lower quality solutions, but finishes faster.
Example: 'PercentNeighbors',0.01
Method for choosing initial cluster medoid positions, specified
as the comma-separated pair consisting of 'Start' and 'plus', 'sample', 'cluster',
or a matrix. This table summarizes the available methods.
| Method | Description |
|---|---|
'plus' (default) | Select k observations from X according
to the k-means++
algorithm for cluster center initialization. |
'sample' | Select k observations from X at
random. |
'cluster' | Perform preliminary clustering phase on a random subsample
(10%) of X. This preliminary phase is itself
initialized using sample, that is, the observations
are selected at random. |
| matrix | A custom k-by-p matrix
of starting locations. In this case, you can pass in [] for
the k input argument, and kmedoids infers k from
the first dimension of the matrix. You can also supply a 3-D array,
implying a value for 'Replicates' from the array’s
third dimension. |
Example: 'Start','sample'
Data Types: char | string | single | double
Output Arguments
Medoid indices, returned as a numeric column vector. idx has
as many rows as X, and each row indicates the
medoid assignment of the corresponding observation.
Cluster medoid locations, returned as a numeric matrix. C is
a k-by-p matrix, where row j is
the medoid of cluster j
Within-cluster sums of point-to-medoid distances, returned as
a numeric column vector. sumd is a k-by1
vector, where element j is the sum of point-to-medoid
distances within cluster j.
Distances from each point to every medoid, returned as a numeric
matrix. D is an n-by-k matrix,
where element (j,m) is the distance
from observation j to medoid m.
Algorithm information, returned as a struct. info contains options used
by the function when executed such as k-medoid clustering
algorithm (algorithm), method used to choose initial
cluster medoid positions (start), distance metric
(distance), number of iterations taken in the best
replicate (iterations) and the replicate number of the
returned results (bestReplicate).
More About
k-medoids clustering is a partitioning method commonly used in domains that require robustness to outlier data, arbitrary distance metrics, or ones for which the mean or median does not have a clear definition.
It is similar to k-means, and the goal of both methods is to divide a set of measurements or observations into k subsets or clusters so that the subsets minimize the sum of distances between a measurement and a center of the measurement’s cluster. In the k-means algorithm, the center of the subset is the mean of measurements in the subset, often called a centroid. In the k-medoids algorithm, the center of the subset is a member of the subset, called a medoid.
The k-medoids algorithm returns medoids which are the actual data points in the data set. This allows you to use the algorithm in situations where the mean of the data does not exist within the data set. This is the main difference between k-medoids and k-means where the centroids returned by k-means may not be within the data set. Hence k-medoids is useful for clustering categorical data where a mean is impossible to define or interpret.
The function kmedoids provides several
iterative algorithms that minimize the sum of distances from each
object to its cluster medoid, over all clusters. One of the algorithms
is called partitioning around medoids (PAM) [1] which
proceeds in two steps.
Build-step: Each of k clusters is associated with a potential medoid. This assignment is performed using a technique specified by the
'Start'name-value pair argument.Swap-step: Within each cluster, each point is tested as a potential medoid by checking if the sum of within-cluster distances gets smaller using that point as the medoid. If so, the point is defined as a new medoid. Every point is then assigned to the cluster with the closest medoid.
The algorithm iterates the build- and swap-steps until the medoids do not change, or other termination criteria are met.
You can control the details of the minimization using several
optional input parameters to kmedoids, including
ones for the initial values of the cluster medoids, and for the maximum
number of iterations. By default, kmedoids uses
the k-means++
algorithm for cluster medoid initialization and the squared
Euclidean metric to determine distances.
References
[1] Kaufman, L., and Rousseeuw, P. J. (2009). Finding Groups in Data: An Introduction to Cluster Analysis. Hoboken, New Jersey: John Wiley & Sons, Inc.
[2] Park, H-S, and Jun, C-H. (2009). A simple and fast algorithm for K-medoids clustering. Expert Systems with Applications. 36, 3336-3341.
[3] Schlimmer,J.S. (1987). Concept Acquisition Through Representational Adjustment (Technical Report 87-19). Doctoral dissertation, Department of Information and Computer Science, University of California, Irvine.
[4] Iba,W., Wogulis,J., and Langley,P. (1988). Trading off Simplicity and Coverage in Incremental Concept Learning. In Proceedings of the 5th International Conference on Machine Learning, 73-79. Ann Arbor, Michigan: Morgan Kaufmann.
[5] Duch W, A.R., and Grabczewski, K. (1996) Extraction of logical rules from training data using backpropagation networks. Proc. of the 1st Online Workshop on Soft Computing, 19-30, pp. 25-30.
[6] Duch, W., Adamczak, R., Grabczewski, K., Ishikawa, M., and Ueda, H. (1997). Extraction of crisp logical rules using constrained backpropagation networks - comparison of two new approaches. Proc. of the European Symposium on Artificial Neural Networks (ESANN'97), Bruge, Belgium 16-18.
[7] Bache, K. and Lichman, M. (2013). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.
Extended Capabilities
To run in parallel, specify the Options name-value argument in the call to
this function and set the UseParallel field of the
options structure to true using
statset:
Options=statset(UseParallel=true)
For more information about parallel computing, see Run MATLAB Functions with Automatic Parallel Support (Parallel Computing Toolbox).
Version History
Introduced in R2014b
See Also
clusterdata | kmeans | linkage | silhouette | pdist | linkage | evalclusters
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)