semanticseg
Semantic image segmentation using deep learning
Syntax
Description
pxds = semanticseg(ds,network)ds, a datastore object.
The function supports parallel computing using multiple MATLAB® workers. You can enable parallel computing using the Computer Vision Toolbox Preferences dialog.
[___] = semanticseg(___,
specifies options using one or more name-value arguments in addition to any
combination of arguments from previous syntaxes. For example,
Name=Value)ExecutionEnvironment="gpu" sets the hardware resource for
processing images to gpu.
Examples
Perform semantic segmentation of a test image and display the results.
Load a pretrained network.
load("triangleSegmentationNetwork")List the network layers.
net.Layers
ans =
9×1 Layer array with layers:
1 'imageinput' Image Input 32×32×1 images with 'zerocenter' normalization
2 'conv_1' 2-D Convolution 64 3×3×1 convolutions with stride [1 1] and padding [1 1 1 1]
3 'relu_1' ReLU ReLU
4 'maxpool' 2-D Max Pooling 2×2 max pooling with stride [2 2] and padding [0 0 0 0]
5 'conv_2' 2-D Convolution 64 3×3×64 convolutions with stride [1 1] and padding [1 1 1 1]
6 'relu_2' ReLU ReLU
7 'transposed-conv' 2-D Transposed Convolution 64 4×4×64 transposed convolutions with stride [2 2] and cropping [1 1 1 1]
8 'conv_3' 2-D Convolution 2 1×1×64 convolutions with stride [1 1] and padding [0 0 0 0]
9 'softmax' Softmax softmax
Read and display the test image.
I = imread("triangleTest.jpg");
imshow(I)
Define the two classes on which the network was trained, then perform semantic image segmentation.
classNames = ["triangle" "background"]; [C,scores] = semanticseg(I,net,Classes=classNames,MiniBatchSize=32);
Overlay segmentation results on the image and display the results.
B = labeloverlay(I,C); imshow(B)

Display the classification confidence scores.
imagesc(scores)
axis square
colorbar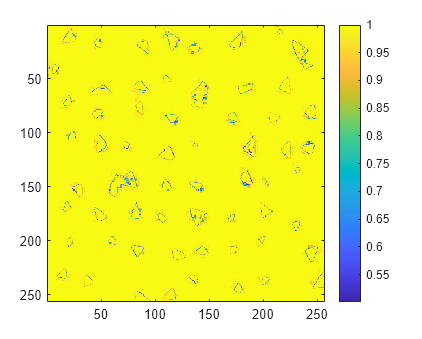
Create a binary mask with only the triangles.
BW = C=="triangle";
imshow(BW)
Run semantic segmentation on a test set of images and compare the results against ground truth data.
Load a pretrained network.
data = load("triangleSegmentationNetwork");
net = data.net;Load test images using imageDatastore.
dataDir = fullfile(toolboxdir("vision"),"visiondata","triangleImages"); testImageDir = fullfile(dataDir,"testImages"); imds = imageDatastore(testImageDir)
imds =
ImageDatastore with properties:
Files: {
' .../toolbox/vision/visiondata/triangleImages/testImages/image_001.jpg';
' .../toolbox/vision/visiondata/triangleImages/testImages/image_002.jpg';
' .../toolbox/vision/visiondata/triangleImages/testImages/image_003.jpg'
... and 97 more
}
Folders: {
' .../runnable/matlab/toolbox/vision/visiondata/triangleImages/testImages'
}
AlternateFileSystemRoots: {}
ReadSize: 1
Labels: {}
SupportedOutputFormats: ["png" "jpg" "jpeg" "tif" "tiff"]
DefaultOutputFormat: "png"
ReadFcn: @readDatastoreImage
Load ground truth test labels.
testLabelDir = fullfile(dataDir,"testLabels"); classNames = ["triangle" "background"]; pixelLabelID = [255 0]; pxdsTruth = pixelLabelDatastore(testLabelDir,classNames,pixelLabelID);
Run semantic segmentation on all of the test images with a batch size of 4. You can increase the batch size to increase throughput based on your systems memory resources.
pxdsResults = semanticseg(imds,net,Classes=classNames, ...
MiniBatchSize=4,WriteLocation=tempdir);Running semantic segmentation network ------------------------------------- * Processed 100 images.
Compare the results against the ground truth.
metrics = evaluateSemanticSegmentation(pxdsResults,pxdsTruth)
Evaluating semantic segmentation results
----------------------------------------
* Selected metrics: global accuracy, class accuracy, IoU, weighted IoU, BF score.
* Processed 100 images.
* Finalizing... Done.
* Data set metrics:
GlobalAccuracy MeanAccuracy MeanIoU WeightedIoU MeanBFScore
______________ ____________ _______ ___________ ___________
0.99074 0.99183 0.91118 0.98299 0.80563
metrics =
semanticSegmentationMetrics with properties:
ConfusionMatrix: [2×2 table]
NormalizedConfusionMatrix: [2×2 table]
DataSetMetrics: [1×5 table]
ClassMetrics: [2×3 table]
ImageMetrics: [100×5 table]
This example shows how to train a semantic segmentation network using dilated convolutions.
A semantic segmentation network classifies every pixel in an image, resulting in an image that is segmented by class. Applications for semantic segmentation include road segmentation for autonomous driving and cancer cell segmentation for medical diagnosis. To learn more, see Get Started with Semantic Segmentation Using Deep Learning.
Semantic segmentation networks like Deeplab v3+ [1] make extensive use of dilated convolutions (also known as atrous convolutions) because they can increase the receptive field of the layer (the area of the input which the layers can see) without increasing the number of parameters or computations.
Load Training Data
The example uses a simple dataset of 32-by-32 triangle images for illustration purposes. The dataset includes accompanying pixel label ground truth data. Load the training data using an imageDatastore and a pixelLabelDatastore.
dataFolder = fullfile(toolboxdir("vision"),"visiondata","triangleImages"); imageFolderTrain = fullfile(dataFolder,"trainingImages"); labelFolderTrain = fullfile(dataFolder,"trainingLabels");
Create an imageDatastore for the images.
imdsTrain = imageDatastore(imageFolderTrain);
Create a pixelLabelDatastore for the ground truth pixel labels.
classNames = ["triangle" "background"]; labels = [255 0]; pxdsTrain = pixelLabelDatastore(labelFolderTrain,classNames,labels)
pxdsTrain =
PixelLabelDatastore with properties:
Files: {200×1 cell}
ClassNames: {2×1 cell}
ReadSize: 1
ReadFcn: @readDatastoreImage
AlternateFileSystemRoots: {}
Create Semantic Segmentation Network
This example uses a simple semantic segmentation network based on dilated convolutions.
Create a data source for training data and get the pixel counts for each label.
ds = combine(imdsTrain,pxdsTrain); tbl = countEachLabel(pxdsTrain)
tbl=2×3 table
Name PixelCount ImagePixelCount
______________ __________ _______________
{'triangle' } 10326 2.048e+05
{'background'} 1.9447e+05 2.048e+05
The majority of pixel labels are for background. This class imbalance biases the learning process in favor of the dominant class. To fix this, use class weighting to balance the classes. You can use several methods to compute class weights. One common method is inverse frequency weighting where the class weights are the inverse of the class frequencies. This method increases the weight given to under represented classes. Calculate the class weights using inverse frequency weighting.
numberPixels = sum(tbl.PixelCount);
frequency = tbl.PixelCount / numberPixels;
classWeights = dlarray(1 ./ frequency,"C");Create a network for pixel classification by using an image input layer with an input size corresponding to the size of the input images. Next, specify three blocks of convolution, batch normalization, and ReLU layers. For each convolutional layer, specify 32 3-by-3 filters with increasing dilation factors and pad the inputs so they are the same size as the outputs by setting the Padding name-value argument as "same". To classify the pixels, include a convolutional layer with K 1-by-1 convolutions, where K is the number of classes, followed by a softmax layer. The classification of pixels is done with a custom model loss within the built-in trainer, trainnet.
inputSize = [32 32 1];
filterSize = 3;
numFilters = 32;
numClasses = numel(classNames);
layers = [
imageInputLayer(inputSize)
convolution2dLayer(filterSize,numFilters,DilationFactor=1,Padding="same")
batchNormalizationLayer
reluLayer
convolution2dLayer(filterSize,numFilters,DilationFactor=2,Padding="same")
batchNormalizationLayer
reluLayer
convolution2dLayer(filterSize,numFilters,DilationFactor=4,Padding="same")
batchNormalizationLayer
reluLayer
convolution2dLayer(1,numClasses)
softmaxLayer];Model Loss Function
The semantic segmentation network can be trained using different loss functions. The built-in trainer trainnet (Deep Learning Toolbox) supports custom loss functions as well as some standard loss functions such as "crossentropy" and "mse". A custom loss function manually computes the loss for each batch of training data by comparing the network's predictions to the actual ground truth or target values. Custom loss functions use a function handle with the function syntax loss = f(Y1,...,Yn,T1,...,Tm), where Y1,...,Yn are dlarray objects that correspond to the n network predictions and T1,...,Tm are dlarray objects that correspond to the m targets.
This example enables you to select from two different loss functions that account for the class imbalance seen in the data. These loss functions are:
Weighted cross-entropy loss, which uses the
crossentropy(Deep Learning Toolbox) function. Weighted cross-entropy loss gives stronger favor to the underrepresented class by scaling the error of that class during training.A custom loss function called
tverskyLossthat calculates the Tversky loss [2]. Tversky loss is more specialized loss for class imbalance.
The Tversky loss is based on the Tversky index for measuring overlap between two segmented images. The Tversky index between one image and the corresponding ground truth is given by
corresponds to the class and corresponds to not being in class .
is the number of elements along the first two dimensions of .
and are weighting factors that control the contribution that false positives and false negatives for each class make to the loss.
The loss over the number of classes is given by
Select the loss function to use during training.
lossFunction =  "tverskyLoss"
"tverskyLoss"lossFunction = "tverskyLoss"
if strcmp(lossFunction,"tverskyLoss") % Declare Tversky loss weighting coefficients for false positives and % false negatives. These coefficients are set and passed to the % training loss function using trainnet. alpha = 0.7; beta = 0.3; lossFcn = @(Y,T) tverskyLoss(Y,T,alpha,beta); else % Use weighted cross-entropy loss during training. lossFcn = @(Y,T) crossentropy(Y,T,classWeights,NormalizationFactor="all-elements"); end
Train Network
Specify the training options.
options = trainingOptions("sgdm",... MaxEpochs=100,... MiniBatchSize= 64,... InitialLearnRate=1e-2,... Verbose=false);
Train the network using trainnet (Deep Learning Toolbox). Specify the loss as the loss function lossFcn.
net = trainnet(ds,layers,lossFcn,options);
Test Network
Load the test data. Create an imageDatastore for the images. Create a pixelLabelDatastore for the ground truth pixel labels.
imageFolderTest = fullfile(dataFolder,"testImages"); imdsTest = imageDatastore(imageFolderTest); labelFolderTest = fullfile(dataFolder,"testLabels"); pxdsTest = pixelLabelDatastore(labelFolderTest,classNames,labels);
Make predictions using the test data and trained network.
pxdsPred = semanticseg(imdsTest,net,... Classes=classNames,... MiniBatchSize=32,... WriteLocation=tempdir);
Running semantic segmentation network ------------------------------------- * Processed 100 images.
Evaluate the prediction accuracy using evaluateSemanticSegmentation.
metrics = evaluateSemanticSegmentation(pxdsPred,pxdsTest);
Evaluating semantic segmentation results
----------------------------------------
* Selected metrics: global accuracy, class accuracy, IoU, weighted IoU, BF score.
* Processed 100 images.
* Finalizing... Done.
* Data set metrics:
GlobalAccuracy MeanAccuracy MeanIoU WeightedIoU MeanBFScore
______________ ____________ _______ ___________ ___________
0.99674 0.98562 0.96447 0.99362 0.92831
Segment New Image
Read the test image triangleTest.jpg and segment the test image using semanticseg. Display the results using labeloverlay.
imgTest = imread("triangleTest.jpg");
[C,scores] = semanticseg(imgTest,net,classes=classNames);
B = labeloverlay(imgTest,C);
montage({imgTest,B})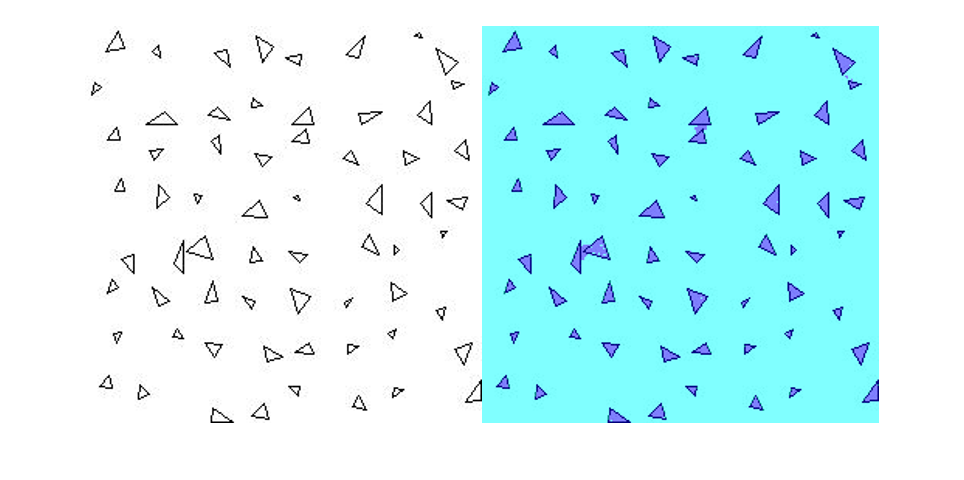
Supporting Functions
function loss = tverskyLoss(Y,T,alpha,beta) % loss = tverskyLoss(Y,T,alpha,beta) returns the Tversky loss % between the predictions Y and the training targets T. Pcnot = 1-Y; Gcnot = 1-T; TP = sum(sum(Y.*T,1),2); FP = sum(sum(Y.*Gcnot,1),2); FN = sum(sum(Pcnot.*T,1),2); epsilon = 1e-8; numer = TP + epsilon; denom = TP + alpha*FP + beta*FN + epsilon; % Compute tversky index. lossTIc = 1 - numer./denom; lossTI = sum(lossTIc,3); % Return average Tversky index loss. N = size(Y,4); loss = sum(lossTI)/N; end
References
[1] Chen, Liang-Chieh et al. “Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation.” ECCV (2018).
[2] Salehi, Seyed Sadegh Mohseni, Deniz Erdogmus, and Ali Gholipour. "Tversky loss function for image segmentation using 3D fully convolutional deep networks." International Workshop on Machine Learning in Medical Imaging. Springer, Cham, 2017.
Input Arguments
Input image, specified as one of the following.
| Image Type | Data Format |
|---|---|
| Single 2-D grayscale image | 2-D matrix of size H-by-W |
| Single 2-D color image or 2-D multispectral image | 3-D array of size H-by-W-by-C. The number of color channels C is 3 for color images. |
| Series of P 2-D images | 4-D array of size H-by-W-by-C-by-P. The number of color channels C is 1 for grayscale images and 3 for color images. |
| Single 3-D grayscale image with depth D | 3-D array of size H-by-W-by-D |
| Single 3-D color image or 3-D multispectral image | 4-D array of size H-by-W-by-D-by-C. The number of color channels C is 3 for color images. |
| Series of P 3-D images | 5-D array of size H-by-W-by-D-by-C-by-P |
The input image can also be a gpuArray (Parallel Computing Toolbox) containing one of
the preceding image types (requires Parallel Computing Toolbox™).
Data Types: uint8 | uint16 | int16 | double | single | logical
Network, specified as a dlnetwork (Deep Learning Toolbox) or taylorPrunableNetwork (Deep Learning Toolbox) object.
Region of interest, specified as one of the following.
| Image Type | ROI Format |
|---|---|
| 2-D image | 4-element vector of the form [x,y,width,height] |
| 3-D image | 6-element vector of the form [x,y,z,width,height,depth] |
The vector defines a rectangular or cuboidal region of
interest fully contained in the input image. Image pixels outside the region
of interest are assigned the <undefined> categorical
label. If the input image consists of a series of images, then
semanticseg applies the same
roi to all images in the series.
Collection of images, specified as a datastore. The read function of the datastore must return a numeric array,
cell array, or table. For cell arrays or tables with multiple columns, the
function processes only the first column.
For more information, see Datastores for Deep Learning (Deep Learning Toolbox).
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: ExecutionEnvironment="gpu" sets the hardware resource
for processing images to "gpu".
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Returned segmentation type, specified as
"categorical", "double", or
"uint8". When you specify
"double" or "uint8", the
function returns the segmentation results as a label array containing
label IDs. The IDs are integer values that correspond to the class names
defined in the classification layer used in the input network.
You cannot use the OutputType property with an
ImageDatastore object
input.
Group of images, specified as an integer. Images are grouped and
processed together as a batch. Batches are used for processing a large
collection of images and they improve computational efficiency.
Increasing the 'MiniBatchSize' value increases the
efficiency, but it also takes up more memory.
Hardware resource for processing images with a network, specified as
"auto", "gpu", or
"cpu".
ExecutionEnvironment | Description |
|---|---|
"auto" | Use a GPU if available. Otherwise, use the CPU. The use of GPU requires Parallel Computing Toolbox and a CUDA® enabled NVIDIA® GPU. For information about the supported compute capabilities, see GPU Computing Requirements (Parallel Computing Toolbox). |
"gpu" | Use the GPU. If a suitable GPU is not available, the function returns an error message. |
"cpu" | Use the CPU. |
Performance optimization, specified as "auto",
"mex", or
"none".
Acceleration | Description |
|---|---|
"auto" | Automatically apply a number of optimizations suitable for the input network and hardware resource. |
"mex" | Compile and execute a MEX function. This option is available when using a GPU only. You must also have a C/C++ compiler installed. For setup instructions, see Set Up Compiler (GPU Coder). |
"none" | Disable all acceleration. |
The default option is "auto". If you use the
"auto" option, then MATLAB does not ever generate a MEX function.
Using the Acceleration name-value argument options
"auto" and "mex" can offer
performance benefits, but at the expense of an increased initial run
time. Subsequent calls with compatible parameters are faster. Use
performance optimization when you plan to call the function multiple
times using new input data.
The "mex" option generates and executes a MEX
function based on the network and parameters used in the function call.
You can have several MEX functions associated with a single network at
one time. Clearing the network variable also clears any MEX functions
associated with that network.
The "mex" option is only available when you are
using a GPU. Using a GPU
requires Parallel Computing Toolbox and a CUDA enabled NVIDIA GPU. For information about the supported
compute capabilities, see GPU Computing Requirements (Parallel Computing Toolbox).
If Parallel Computing Toolbox or a suitable GPU is not available, then the function
returns an error.
"mex" acceleration does not support all layers. For
a list of supported layers, see Supported Layers (GPU Coder).
Classes into which pixels or voxels are classified, specified as
"auto", a cell array of character vectors, a
string vector, or a categorical vector. If the value is a categorical
vector Y, then the elements of the vector are sorted
and ordered according to categories(Y).
If the network is a dlnetwork (Deep Learning Toolbox) object,
then the number of classes specified by 'Classes'
must match the number of channels in the output of the network
predictions. By default, when 'Classes' has the
value "auto", the classes are numbered from 1 through
C, where C is the number of
channels in the output layer of the network.
If the network is a SeriesNetwork (Deep Learning Toolbox) or DAGNetwork (Deep Learning Toolbox) object, then the number of classes specified by
the Classes name-value argument must match the
number of classes in the classification output layer. By default, when
Classes has the value
"auto", the classes are automatically set using the
classification output layer.
Folder location, specified as pwd (your current
working folder), a string scalar, or a character vector. The specified
folder must exist and have write permissions.
This property applies only when using a datastore that can process images.
Prefix applied to output filenames, specified as a string scalar or character vector. The image files are named as follows:
<, whereprefix>_<N>.pngNimds.Files(N).
This property applies only when using a datastore that can process images.
Output folder name for segmentation results, specified as a string
scalar or a character vector. This folder is in the location specified
by the value of the WriteLocation name-value
argument.
If the output folder already exists, the function creates a new folder
with the string "_1" appended to the end of the name.
Set OutputFoldername to "" to
write all the results to the folder specified by
WriteLocation.
Suffix to add to the output image filename, specified as a string scalar or a character vector. The function appends the specified suffix to the output filename as:
<, whereprefix>_<N><suffix>.pngNimds.Files(N).
If you do not specify the suffix, the function uses the
input filenames as the output file suffixes. The function extracts the
input filenames from the info output of the
read object function of the datastore. When the
datastore does not provide the filename, the function does not add a
suffix.
Display progress information, specified as a logical
0 (false) or
1 (true). Specify
Verbose as true to display
progress information. This property applies only when using a datastore
that can process images.
Output Arguments
Categorical labels, returned as a categorical array. The categorical array
relates a label to each pixel or voxel in the input image. The images
returned by readall(datastore) have a one-to-one
correspondence with the categorical matrices returned by readall(pixelLabelDatastore). The elements of the label array
correspond to the pixel or voxel elements of the input image. If you select
an ROI, then the labels are limited to the area within the ROI. Image pixels
and voxels outside the region of interest are assigned the
<undefined> categorical label.
| Image Type | Categorical Label Format |
|---|---|
| Single 2-D image | 2-D matrix of size
H-by-W.
Element
C(i,j)
is the categorical label assigned to the pixel
I(i,j). |
| Series of P 2-D images | 3-D array of size
H-by-W-by-P.
Element
C(i,j,p)
is the categorical label assigned to the pixel
I(i,j,p). |
| Single 3-D image | 3-D array of size
H-by-W-by-D.
Element
C(i,j,k)
is the categorical label assigned to the voxel
I(i,j,k). |
| Series of P 3-D images | 4-D array of size
H-by-W-by-D-by-P.
Element
C(i,j,k,p)
is the categorical label assigned to the voxel
I(i,j,k,p). |
Confidence scores for each categorical label in C,
returned as an array of values between 0 and
1. The scores represents the confidence in the
predicted labels C. Higher score values indicate a
higher confidence in the predicted label.
| Image Type | Score Format |
|---|---|
| Single 2-D image | 2-D matrix of size
H-by-W.
Element
score(i,j)
is the classification score of the pixel
I(i,j). |
| Series of P 2-D images | 3-D array of size
H-by-W-by-P.
Element
score(i,j,p)
is the classification score of the pixel
I(i,j,p). |
| Single 3-D image | 3-D array of size
H-by-W-by-D.
Element
score(i,j,k)
is the classification score of the voxel
I(i,j,k). |
| Series of P 3-D images | 4-D array of size
H-by-W-by-D-by-P.
Element
score(i,j,k,p)
is the classification score of the voxel
I(i,j,k,p). |
Scores for all label categories that the input network can classify, returned as a numeric array. The format of the array is described in the following table. L represents the total number of label categories.
| Image Type | All Scores Format |
|---|---|
| Single 2-D image | 3-D array of size
H-by-W-by-L.
Element
allScores(i,j,q)
is the score of the qth label at the
pixel
I(i,j). |
| Series of P 2-D images | 4-D array of size
H-by-W-by-L-by-P.
Element
allScores(i,j,q,p)
is the score of the qth label at the
pixel
I(i,j,p). |
| Single 3-D image | 4-D array of size
H-by-W-by-D-by-L.
Element
allScores(i,j,k,q)
is the score of the qth label at the
voxel
I(i,j,k). |
| Series of P 3-D images | 5-D array of size
H-by-W-by-D-by-L-by-P.
Element
allScores(i,j,k,q,p)
is the score of the qth label at the
voxel
I(i,j,k,p). |
Semantic segmentation results, returned as a pixelLabelDatastore object. The object contains the semantic
segmentation results for all the images contained in the
ds input object. The result for each image is saved
as separate uint8 label matrices of PNG images. You can
use read(pxds) to return the categorical
labels assigned to the images in ds.
The images in the output of readall(ds) have a one-to-one
correspondence with the categorical matrices in the output of readall(pxds).
Extended Capabilities
Usage notes and limitations:
To prepare a
semanticSegobject for code generation, usecoder.loadDeepLearningNetwork.For code generation, the
semanticSegobject does not supportimageDatastoreand 3-D images as input.For code generation,
ExcecutionEnvironmentandAccelerationname value pairs are ignored with compile time warnings.For code generation, only
OutputType,MiniBatchSize, andClassesname value arguments are supported. These name value arguments must be compile time constants.For code generation,
Classesname value argument does not support string array as input.For code generation,
pxdsoutput argument is not supported.
Usage notes and limitations:
To prepare a
semanticSegobject for code generation, usecoder.loadDeepLearningNetwork.For code generation, the
semanticSegobject does not supportimageDatastoreand 3-D images as input.For code generation,
ExcecutionEnvironmentandAccelerationname value pairs are ignored with compile time warnings.For code generation, only
OutputType,MiniBatchSize, andClassesname value arguments are supported. These name value arguments must be compile time constants.For code generation,
Classesname value argument does not support string array as input.For code generation,
pxdsoutput argument is not supported.
To run in parallel, set 'UseParallel' to true or enable
this by default using the Computer Vision Toolbox™ preferences.
For more information, see Parallel Computing Toolbox Support.
Version History
Introduced in R2017bStarting in R2024a, DAGNetwork (Deep Learning Toolbox) and SeriesNetwork (Deep Learning Toolbox) objects are not recommended. Instead, specify the
semantic segmentation network as a dlnetwork (Deep Learning Toolbox) object.
There are no plans to remove support for DAGNetwork and
SeriesNetwork objects. However, dlnetwork
objects have these advantages:
dlnetworkobjects support a wider range of network architectures which you can then easily train using thetrainnet(Deep Learning Toolbox) function or import from external platforms.dlnetworkobjects provide more flexibility. They have wider support with current and upcoming Deep Learning Toolbox™ functionality.dlnetworkobjects provide a unified data type that supports network building, prediction, built-in training, compression, and custom training loops.dlnetworktraining and prediction is typically faster thanDAGNetworkandSeriesNetworktraining and prediction.
The semanticseg function now writes output files to the
folder specified by the WriteLocation and
OutputFolderName name-value arguments as
<WriteLocation>/<OutputFolderName>. Prior to R2023a,
the function wrote output files directly into the location specified by
WriteLocation. To get the same results as in previous
releases, set the OutputFolderName name-value argument to
"".
See Also
Apps
Functions
trainnet(Deep Learning Toolbox) |labeloverlay|evaluateSemanticSegmentation
Objects
ImageDatastore|pixelLabelDatastore|dlnetwork(Deep Learning Toolbox)
Topics
- Get Started with Semantic Segmentation Using Deep Learning
- Deep Learning in MATLAB (Deep Learning Toolbox)
- Datastores for Deep Learning (Deep Learning Toolbox)
External Websites
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)