使用深度学习对多光谱图像进行语义分割
此示例说明如何使用 U-Net 对包含七个通道的多光谱图像执行语义分割。
语义分割涉及用类标注图像中的每个像素。语义分割的一个应用是跟踪森林采伐,即森林植被随时间的变化。环保机构需要跟踪森林采伐,以评估和量化某个地区的环境和生态健康。
基于深度学习的语义分割可以通过高分辨率航拍照片精确测量植被覆盖度。为了提高分类准确度,一些数据集包含多光谱图像,这些图像提供关于每个像素的附加信息。例如,Hamlin Beach 国家公园数据集用三个近红外通道为彩色图像补充信息,以提供更清晰的分类。
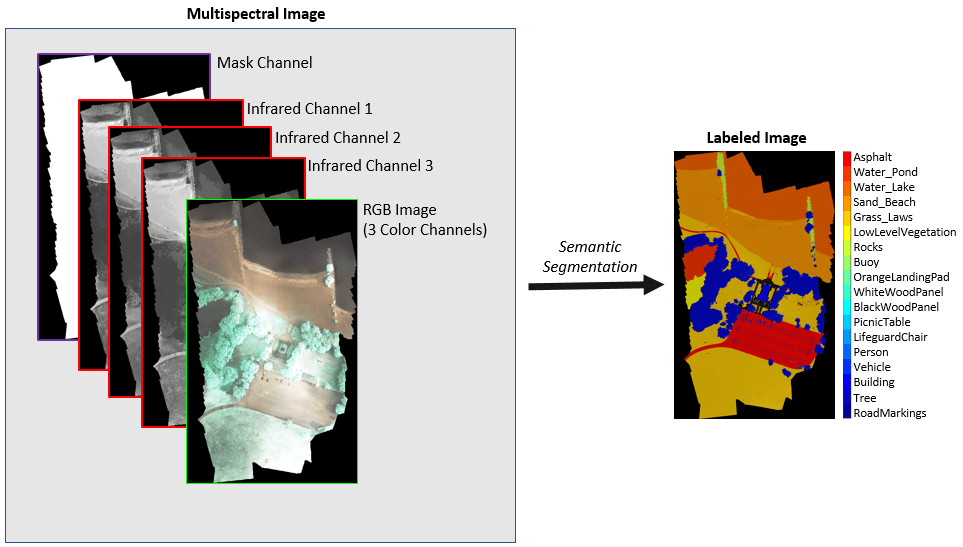
此示例首先说明如何使用预训练的 U-Net 执行语义分割,然后使用分割结果来计算植被覆盖率。然后,您可以选择使用基于补片的训练方法基于 Hamlin Beach State Park 数据集训练 U-Net 网络。
下载预训练的 U-Net
指定 dataDir 作为经过训练的网络和数据集的期望位置。
dataDir = fullfile(tempdir,"rit18_data");下载预训练的 U-Net 网络。
trainedNet_url = "https://ssd.mathworks.com/supportfiles/"+ ... "vision/data/trainedMultispectralUnetModel_v2.zip"; downloadTrainedNetwork(trainedNet_url,dataDir); load(fullfile(dataDir,"trainedMultispectralUnetModel", ... "trainedMultispectralUnetModel_v2.mat"));
下载数据集
此示例使用高分辨率多光谱数据集来训练网络 [1]。图像集是用无人机在纽约 Hamlin Beach 州立公园拍摄的。数据包含已标注的训练集、验证集和测试集,有 18 个对象类标签。数据文件的大小为 3.0 GB。
使用 downloadHamlinBeachMSIData 辅助函数下载数据集的 MAT 文件版本。此函数作为支持文件包含在本示例中。
downloadHamlinBeachMSIData(dataDir);
加载数据集。
load(fullfile(dataDir,"rit18_data.mat")); whos train_data val_data test_data
Name Size Bytes Class Attributes test_data 7x12446x7654 1333663576 uint16 train_data 7x9393x5642 741934284 uint16 val_data 7x8833x6918 855493716 uint16
多光谱图像数据排列为通道数×宽度×高度数组。但是,在 MATLAB® 中,多通道图像排列为宽度×高度×通道数数组。要重构数据以使通道处于第三个维度中,请使用 permute 函数。
train_data = permute(train_data,[2 3 1]); val_data = permute(val_data,[2 3 1]); test_data = permute(test_data,[2 3 1]);
确认数据结构正确。
whos train_data val_data test_data
Name Size Bytes Class Attributes test_data 12446x7654x7 1333663576 uint16 train_data 9393x5642x7 741934284 uint16 val_data 8833x6918x7 855493716 uint16
可视化多光谱数据
显示以纳米为单位的每个频谱段的中心。
disp(band_centers)
490 550 680 720 800 900
在此数据集中,RGB 颜色通道分别是第 3 个、第 2 个和第 1 个图像通道。将训练、验证和测试图像的 RGB 分量显示为蒙太奇形式。要使图像在屏幕上看起来更亮,请使用 histeq 函数执行直方图均衡化处理。
rgbTrain = histeq(train_data(:,:,[3 2 1]));
rgbVal = histeq(val_data(:,:,[3 2 1]));
rgbTest = histeq(test_data(:,:,[3 2 1]));
montage({rgbTrain,rgbVal,rgbTest},BorderSize=10,BackgroundColor="white")
title("RGB Component of Training, Validation, and Test Image (Left to Right)")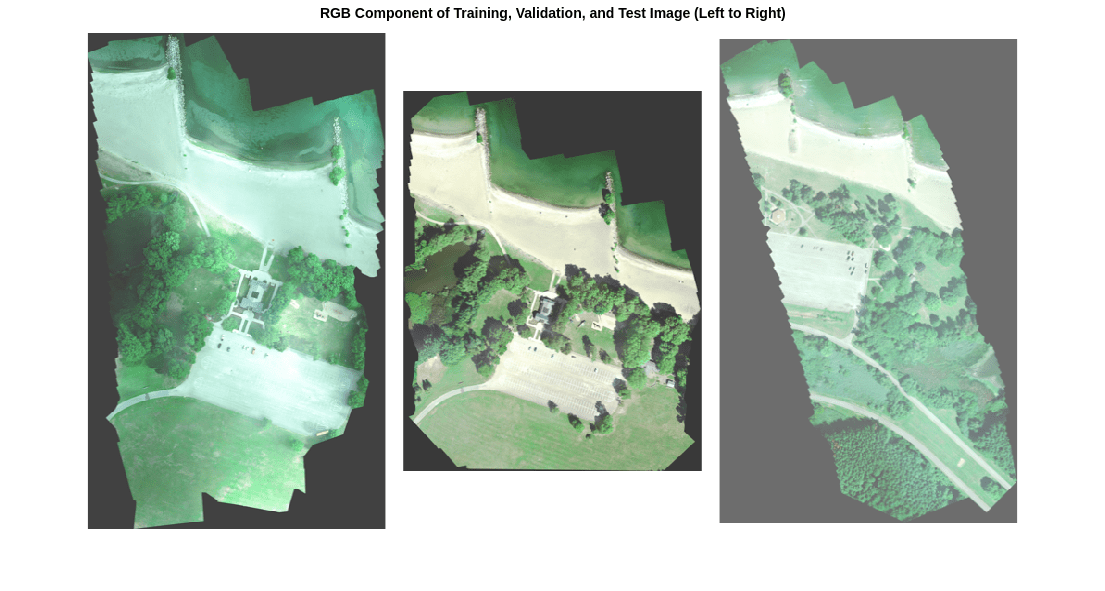
数据的第 4 个、第 5 个和第 6 个通道对应于近红外波段。均衡化处理训练图像的这三个通道的直方图,然后将这些通道以蒙太奇方式显示。这些通道根据其热能特征突出显示图像的不同分量。例如,第 4 个通道中的树比其他两个红外通道中的树更暗。
ir4Train = histeq(train_data(:,:,4));
ir5Train = histeq(train_data(:,:,5));
ir6Train = histeq(train_data(:,:,6));
montage({ir4Train,ir5Train,ir6Train},BorderSize=10,BackgroundColor="white")
title("Infrared Channels 4, 5, and 6 (Left to Right) of Training Image ")
数据的第 7 个通道是表示有效分割区域的二值掩膜。显示训练、验证和测试图像的掩膜。
maskTrain = train_data(:,:,7);
maskVal = val_data(:,:,7);
maskTest = test_data(:,:,7);
montage({maskTrain,maskVal,maskTest},BorderSize=10,BackgroundColor="white")
title("Mask of Training, Validation, and Test Image (Left to Right)")
可视化真实值标签
已标注的图像包含用于分割的真实值数据,每个像素分配给 18 个类之一。获取具有对应 ID 的类的列表。
disp(classes)
0. Other Class/Image Border 1. Road Markings 2. Tree 3. Building 4. Vehicle (Car, Truck, or Bus) 5. Person 6. Lifeguard Chair 7. Picnic Table 8. Black Wood Panel 9. White Wood Panel 10. Orange Landing Pad 11. Water Buoy 12. Rocks 13. Other Vegetation 14. Grass 15. Sand 16. Water (Lake) 17. Water (Pond) 18. Asphalt (Parking Lot/Walkway)
此示例旨在将图像分为两类:植被和非植被。定义目标类名称。
classNames = ["NotVegetation" "Vegetation"];
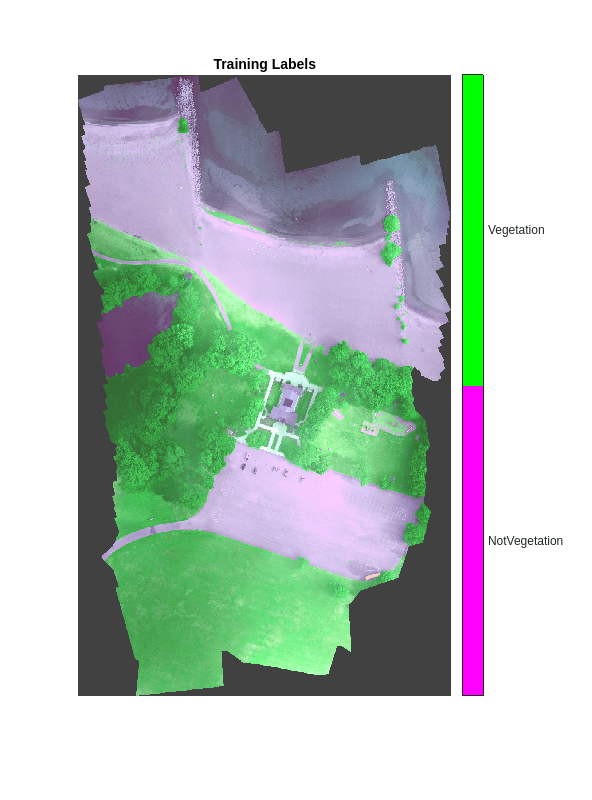
将 18 个原始类分成两个目标类,用于训练和验证数据。Vegetation 是原始类 Tree、Other Vegetation 和 Grass 的组合,这些原始类的 ID 为 2、13 和 14。类 ID 为 0 的原始类 Other Class/Image Border 属于背景类。所有其他原始类都属于目标标签 NotVegetation。
vegetationClassIDs = [2 13 14]; nonvegetationClassIDs = setdiff(1:length(classes),vegetationClassIDs); labelsTrain = zeros(size(train_labels),"uint8"); labelsTrain(ismember(train_labels,nonvegetationClassIDs)) = 1; labelsTrain(ismember(train_labels,vegetationClassIDs)) = 2; labelsVal = zeros(size(val_labels),"uint8"); labelsVal(ismember(val_labels,nonvegetationClassIDs)) = 1; labelsVal(ismember(val_labels,vegetationClassIDs)) = 2;
将真实值验证标签保存为 PNG 文件。此示例将使用此文件来计算准确度度量。
imwrite(labelsVal,"gtruth.png");在经过直方图均衡化处理的 RGB 训练图像上叠加标签。向图像添加颜色栏。
cmap = [1 0 1;0 1 0]; B = labeloverlay(rgbTrain,labelsTrain,Transparency=0.8,Colormap=cmap); imshow(B,cmap) title("Training Labels") numClasses = numel(classNames); ticks = 1/(numClasses*2):1/numClasses:1; colorbar(TickLabels=cellstr(classNames), ... Ticks=ticks,TickLength=0,TickLabelInterpreter="none");
对测试图像执行语义分割
图像的大小导致无法一次性分割整个图像。在这种情况下,请改用分块图像方法分割图像。此方法可以扩展到处理超大型文件,因为它一次只加载和处理一个数据块。
使用 blockedImage 函数创建一个包含测试数据的六个频谱通道的分块图像。
patchSize = [1024 1024]; bimTest = blockedImage(test_data(:,:,1:6),BlockSize=patchSize);
使用 semanticseg 函数对一个数据块进行分割。使用 apply 函数对分块图像中的所有块调用 sematicseg 函数。
bimSeg = apply(bimTest,@(bs)semanticseg(bs.Data,net,Outputtype="uint8"), ... PadPartialBlocks=true,PadMethod=0);
使用 gather 函数将所有分割的分块组合成工作区中的单个图像。
segmentedImage = gather(bimSeg);
为了只提取分割的有效部分,将分割的图像乘以测试数据的掩膜通道。
segmentedImage = segmentedImage .* uint8(maskTest~=0);
imshow(segmentedImage,[])
title("Segmented Image")
语义分割的输出含有噪声。执行图像后处理以去除噪声和杂散像素。使用 medfilt2 函数从分割中去除椒盐噪声。显示去除噪声后的分割图像。
segmentedImage = medfilt2(segmentedImage,[7 7]);
imshow(segmentedImage,[]);
title("Segmented Image with Noise Removed")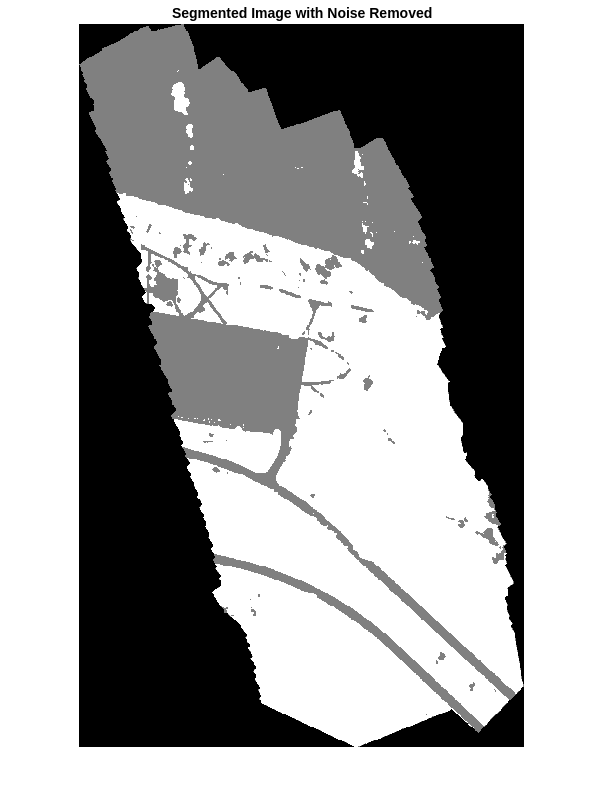
在经过直方图均衡化处理的 RGB 验证图像上叠加分割后的图像。
B = labeloverlay(rgbTest,segmentedImage,Transparency=0.8,Colormap=cmap); imshow(B,cmap) title("Labeled Segmented Image") colorbar(TickLabels=cellstr(classNames), ... Ticks=ticks,TickLength=0,TickLabelInterpreter="none");
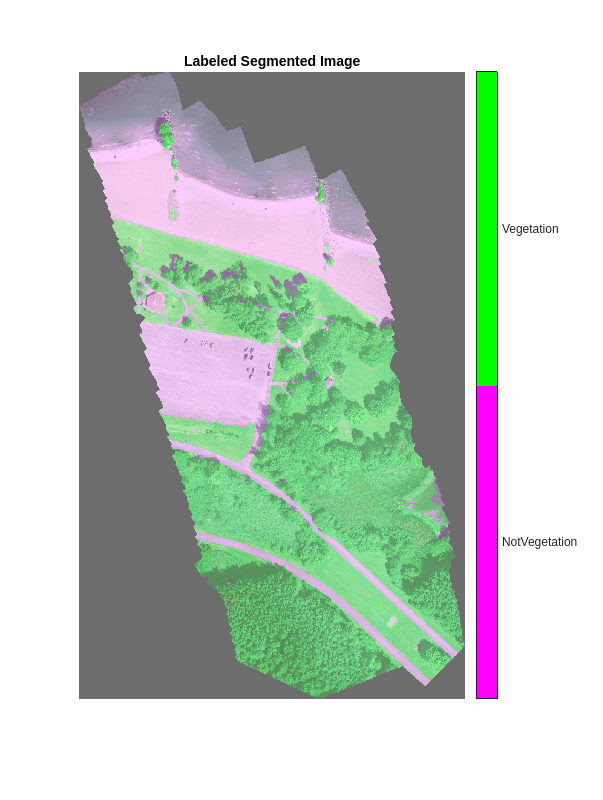
计算植被覆盖率
语义分割结果可用于解决相关生态学问题。例如,植被覆盖的陆地面积百分比是多少?要回答此问题,请找出分割的测试图像中标注为植被的像素数。还可以通过对分割图像中的非零像素进行计数来确定 ROI 中的像素总数。
vegetationPixels = ismember(segmentedImage(:),vegetationClassIDs); numVegetationPixels = sum(vegetationPixels(:)); numROIPixels = nnz(segmentedImage);
通过用植被像素数除以 ROI 中的像素数来计算植被覆盖率。
percentVegetationCover = (numVegetationPixels/numROIPixels)*100; disp("The percentage of vegetation cover is "+percentVegetationCover+"%");
The percentage of vegetation cover is 60.7028%
该示例的其余部分说明如何基于 Hamlin Beach 数据集训练 U-Net。
创建用于训练的分块图像数据存储
使用一个分块图像数据存储来向网络馈送训练数据。此数据存储从一个包含真实值图像的图像数据存储和包含像素标签的像素标签数据存储中提取多个对应的补片。
将训练图像、训练标签和掩膜作为分块图像读取。
inputTileSize = [256 256]; bim = blockedImage(train_data(:,:,1:6),BlockSize=inputTileSize); bLabels = blockedImage(labelsTrain,BlockSize=inputTileSize); bmask = blockedImage(maskTrain,BlockSize=inputTileSize);
选择与掩膜重叠的图像数据块。
overlapPct = 0.185; blockOffsets = round(inputTileSize.*overlapPct); bls = selectBlockLocations(bLabels, ... BlockSize=inputTileSize,BlockOffsets=blockOffsets, ... Masks=bmask,InclusionThreshold=0.95);
使用 one-hot 方法对标签进行编码。
labelsTrain1hot = onehotencode(labelsTrain,3,ClassNames=1:2); labelsTrain1hot(isnan(labelsTrain1hot)) = 0; bLabels = blockedImage(labelsTrain1hot,BlockSize=inputTileSize);
使用 blockedImageDatastore 函数将数据写入分块图像数据存储。
bimds = blockedImageDatastore(bim,BlockLocationSet=bls,PadMethod=0); bimdsLabels = blockedImageDatastore(bLabels,BlockLocationSet=bls,PadMethod=0);
基于这两个分块图像数据存储创建一个 CombinedDatastore。
dsTrain = combine(bimds,bimdsLabels);
分块图像数据存储 dsTrain 在一轮训练的每次迭代中向网络提供小批量数据。预览数据存储以探查数据。
preview(dsTrain)
ans=1×2 cell array
{256×256×6 uint16} {256×256×2 double}
创建 U-Net 网络
此示例使用 U-Net 网络的一种变体。U-Net 中的初始卷积层序列与最大池化层交叠,从而逐步降低输入图像的分辨率。这些层后跟一系列使用上采样算子散布的卷积层,从而会连续增加输入图像的分辨率 [2]。U-Net 的名称源于网络可以绘制成形似字母 U 的对称形状。
指定 U-Net 的超参数。输入深度是高光谱通道的数量,即 6。
inputDepth = 6;
使用 unet 函数创建 U-Net 网络。
unetNetwork = unet([inputTileSize inputDepth],numClasses);
定义损失函数
定义名为 lossFcn 的自定义损失函数,用于计算图像的所有非掩膜像素的交叉熵损失。
function loss = lossFcn(Y,T) mask = ~isnan(T); T(isnan(T)) = 0; loss = crossentropy(Y,T,Mask=mask); end
选择训练选项
使用具有动量的随机梯度下降 (SGDM) 优化来训练网络。使用 trainingOptions (Deep Learning Toolbox) 函数指定 SGDM 的超参数设置。要启用梯度裁剪,请将 GradientThreshold 名称-值参量指定为 0.05,并将 GradientThresholdMethod 指定为使用梯度的 L2-范数。
maxEpochs = 70; minibatchSize = 16; options = trainingOptions("sgdm", ... InitialLearnRate=0.05, ... MaxEpochs=maxEpochs, ... MiniBatchSize=minibatchSize, ... L2Regularization=0.001, ... LearnRateDropPeriod=10, ... LearnRateSchedule="piecewise", ... GradientThreshold=0.05, ... Shuffle="every-epoch", ... VerboseFrequency=200);
训练网络
要训练网络,请将以下代码中的 doTraining 变量设置为 true。使用 trainnet (Deep Learning Toolbox) 函数训练模型。将损失指定为自定义损失函数 lossFcn。默认情况下,trainnet 函数使用 GPU(如果有)。在 GPU 上进行训练需要 Parallel Computing Toolbox™ 许可证和受支持的 GPU 设备。有关受支持设备的信息,请参阅GPU 计算要求 (Parallel Computing Toolbox)。否则,trainnet 函数使用 CPU。要指定执行环境,请使用 ExecutionEnvironment 训练选项。
doTraining =false; if doTraining net = trainnet(dsTrain,unetNetwork,@lossFcn,options); modelDateTime = string(datetime("now",Format="yyyy-MM-dd-HH-mm-ss")); save(fullfile(dataDir,"multispectralUnet-"+modelDateTime+".mat"),"net"); end
评估分割准确度
分割验证数据。
使用 blockedImage 函数创建一个包含验证数据的六个频谱通道的分块图像。
bimVal = blockedImage(val_data(:,:,1:6),BlockSize=patchSize);
使用 semanticseg 函数对一个数据块进行分割。使用 apply 函数对分块图像中的所有块调用 sematicseg 函数。
bimSeg = apply(bimVal,@(bs)semanticseg(bs.Data,net,Outputtype="uint8"),... PadPartialBlocks=true,PadMethod=0);
使用 gather 函数将所有分割的分块组合成工作区中的单个图像。
segmentedImage = gather(bimSeg);
将分割的图像保存为一个 PNG 文件。
imwrite(segmentedImage,"results.png");使用 pixelLabelDatastore 函数加载分段结果和真实值标签。
pixelLabelIDs = [1 2]; pxdsResults = pixelLabelDatastore("results.png",classNames,pixelLabelIDs); pxdsTruth = pixelLabelDatastore("gtruth.png",classNames,pixelLabelIDs);
使用 evaluateSemanticSegmentation 函数衡量语义分割的准确度。全局准确度分数表明,正确分类的像素大于 96%。
ssm = evaluateSemanticSegmentation(pxdsResults,pxdsTruth);
Evaluating semantic segmentation results
----------------------------------------
* Selected metrics: global accuracy, class accuracy, IoU, weighted IoU, BF score.
* Processed 1 images.
* Finalizing... Done.
* Data set metrics:
GlobalAccuracy MeanAccuracy MeanIoU WeightedIoU MeanBFScore
______________ ____________ _______ ___________ ___________
0.9694 0.9704 0.9406 0.94064 0.79474
参考资料
[1] Kemker, R., C. Salvaggio, and C. Kanan."High-Resolution Multispectral Dataset for Semantic Segmentation."CoRR, abs/1703.01918. 2017.
[2] Ronneberger, O., P. Fischer, and T. Brox."U-Net:Convolutional Networks for Biomedical Image Segmentation."CoRR, abs/1505.04597. 2015.
[3] Kemker, Ronald, Carl Salvaggio, and Christopher Kanan."Algorithms for Semantic Segmentation of Multispectral Remote Sensing Imagery Using Deep Learning."ISPRS Journal of Photogrammetry and Remote Sensing, Deep Learning RS Data, 145 (November 1, 2018):60-77. https://doi.org/10.1016/j.isprsjprs.2018.04.014.
另请参阅
blockedImage | apply | selectBlockLocations | blockedImageDatastore | semanticseg | evaluateSemanticSegmentation | unet | dlnetwork (Deep Learning Toolbox) | trainingOptions (Deep Learning Toolbox) | trainnet (Deep Learning Toolbox)
主题
- Get Started with Semantic Segmentation Using Deep Learning
- 使用深度学习进行语义分割
- Label Large Images in the Image Labeler
- Datastores for Deep Learning (Deep Learning Toolbox)