Detect Defects Using Tiled Training of EfficientAD Anomaly Detector
This example shows how to detect and localize industrial production defects on chewing gum images by training an EfficientAD anomaly detection network on tiled images.
The EfficientAD model [1] is a one-class anomaly detector that uses features extracted from a lightweight convolutional neural network (CNN) to distinguish normal and anomalous images in real-time. You train the model using only normal (non-anomalous) images. To detect local anomalies, the detector operates on the principle of a student-teacher model, where the teacher is a pretrained deep neural network that understands normal data behavior, and the student is a smaller network that learns to predict the features of normal images output by a teacher model. At inference time, both the student and teacher evaluate image data, and the model detects anomalies using the discrepancy between the student and teacher when the student fails to predict the anomaly features. In addition to structural anomalies, EfficientAD can detect logical anomalies that result from incorrect object orientation by using an autoencoder that is tuned to detect global anomalies. The high accuracy, low latency, and high throughput of EfficientAD enables you to detect and localize defects in large-scale applications that require a real-time or near-real-time response.
In this example, you train an EfficientAD network on tiled normal (non-anomalous) images and perform inference on full-size images. During training, you split the images in the data set into full-resolution segments, or tiles. This process preserves the full spatial resolution in the trained detector and is ideal for the detection of small local defects when the autoencoder is off. Evaluate the results using anomaly detection metrics and visual inspection of anomaly maps.
Download Pretrained EfficientAD Detector
By default, this example downloads a pretrained version of the EfficientAD anomaly detector using the helper function downloadTrainedNetwork. The function is attached to this example as a supporting file. You can use the pretrained network to run the entire example without waiting for training to complete. The fully convolutional architecture of EfficientAD enables tiled training and full-size inference without the need for multiple inference calls per image.
trainedGumDefectDetectorNet_url = "https://ssd.mathworks.com/supportfiles/" + ... "vision/data/trainedGumDefectDetectorEfficientAD.zip"; downloadTrainedNetwork(trainedGumDefectDetectorNet_url,pwd) load("trainedGumDefectDetectorEfficientAD.mat")
Download VisA Data Set
Load the Visual Anomaly (VisA) data set of 10,821 high-resolution color images (9,621 normal and 1,200 anomalous samples) covering 12 different object subsets in 3 domains [2]. Several data subsets contain industrial production images of chewing gum, cashews, fryums, and other food. The anomalous images in the chewing gum test set contain surface defects such as scratches, dents, color spots as well as structural defects such as broken edges.
This example uses the chewing gum data subset. This data subset contains train and test folders, which include the normal training images, and the normal and anomalous test images, respectively.
Specify dataDir as the location of the data set. Download the data set using the downloadVisAData helper function. This function, which is attached to the example as a supporting file, downloads a ZIP file and extracts the data.
dataDir = fullfile(tempdir,"VisA");
downloadVisAData(dataDir)Localize Defects in Image
Read a sample anomalous image with a "bad" label from the data set.
sampleImage = imread(fullfile(dataDir,"VisA", ... "chewinggum","test","bad","000.JPG"));
Visualize the localization of defects by displaying the original chewing gum image with the overlaid predicted per-pixel anomaly score map. Use the anomalyMap function to generate the anomaly score heatmap for the sample image. Display the image, with the heatmap overlaid by using the anomalyMapOverlay function.
anomalyHeatMap = anomalyMap(detector,sampleImage);
heatMapImage = anomalyMapOverlay(sampleImage,anomalyHeatMap);
montage({sampleImage,heatMapImage})
title("Heatmap of Anomalous Image")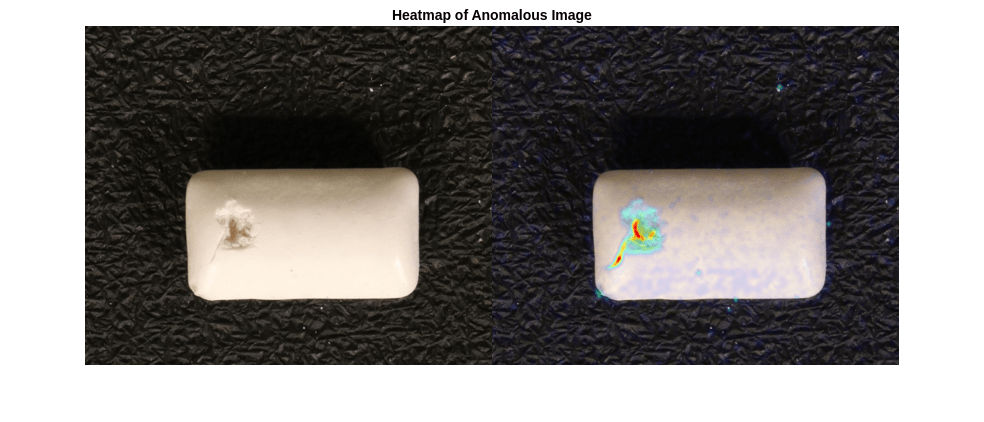
Prepare Data for Training
Create imageDatastore objects that hold the training and test sets, from the train and test folders of the downloaded VisA data set.
exts = {".jpg",".png",".tif"};
dsTrain = imageDatastore(fullfile(dataDir,"VisA","chewinggum","train"),IncludeSubfolders=true,LabelSource="foldernames");
summary(dsTrain.Labels)453×1 categorical
good 453
<undefined> 0
normalLabelStr = "good"; dsTest = imageDatastore(fullfile(dataDir,"VisA","chewinggum","test"),IncludeSubfolders=true,LabelSource="foldernames"); summary(dsTest.Labels)
150×1 categorical
bad 100
good 50
<undefined> 0
Display images of a normal piece of chewing gum and a defective piece of chewing gum from the test data set.
badImage = find(dsTest.Labels=="bad",1); badImage = read(subset(dsTest,badImage)); normalImage = find(dsTest.Labels=="good",1); normalImage = read(subset(dsTest,normalImage)); montage({normalImage,badImage}) title("Test Chewing Gum Images Without (Left) and With (Right) Defects")

Partition Data into Calibration and Test Sets
Prior to partitioning the data, set the global random state to "default" to ensure reproducibility.
rng("default")Use a calibration set to determine the threshold for the classifier. Using separate calibration and test sets avoids information leaking from the test set into the design of the classifier. The classifier labels images with anomaly scores above the threshold as anomalous.
To establish a suitable threshold for the classifier, allocate 50% of the original test set as the calibration set dsCal, which has equal numbers of normal and anomalous images.
[dsCal,dsTest] = splitEachLabel(dsTest,0.5,"randomized");Prepare Data For Training
Split Data into Masked Tiles
Create a blockedImage object from the images in the training set. The blockedImage object represents an image as a collection of smaller tiles, or blocks, of size 256-by-256 pixels. For each block, create a logical mask, in which true (or 1) pixels represent regions of interest that contain meaningful normal data, by specifying the maskObjectsInNormalImages helper function handle to the apply function as the processing function input. Save the blocked image masks normalMasks to the location of the mask folder you create trainNormalMaskDir.
trainNormalMaskDir = fullfile(dataDir,"EAD","chewinggum","train",normalLabelStr+"_masks"); blockSize = [256 256]; if ~isfolder(trainNormalMaskDir) bims = blockedImage(dsTrain.Files); % Find a global threshold from one normal image to segment % chewing gum images. Images in this data set are mostly uniformly % illuminated so the same threshold can be used for all images. im = gather(bims(1)); T = graythresh(im); normalMasks = apply(bims,@(x)maskObjectsInNormalImages(x,T), ... BlockSize=blockSize,OutputLocation=trainNormalMaskDir, ... Adapter=images.blocked.PNGBlocks,UseParallel=false); save(fullfile(trainNormalMaskDir,normalLabelStr+"_masks.mat"),"normalMasks") else load(fullfile(trainNormalMaskDir,normalLabelStr+"_masks.mat"),"normalMasks") end
Select Training Tiles and Write Data to Disk
To accelerate training speed, create an on-disk representation of the blocked image mask data. Select blocked tiles for training that include a normal image of an object with meaningful data by using the selectBlockLocations function. To help the detector learn the background features also present in anomalous images, specify the InclusionThreshold argument as 0.
Write image tiles to disk using the writeall function, and display sample tiles. If you have a Parallel Computing Toolbox™ license, specify the UseParallel argument as true to reduce processing time.
normalBlocksDir = fullfile(dataDir,"EAD","chewinggum","train",normalLabelStr); if ~isfolder(normalBlocksDir) bims = blockedImage(dsTrain.Files); % Select blocks based on the masks computed in the previous step bls = selectBlockLocations(bims,Levels=1,BlockSize=blockSize, ... Mask=normalMasks,ExcludeIncompleteBlocks=true,BlockOffsets=blockSize, ... InclusionThreshold=0,UseParallel=false); bimds = blockedImageDatastore(bims,BlockLocationSet=bls); % Write all tiles to disk writeall(bimds,normalBlocksDir,UseParallel=false) % Display a few blocks bimds.ReadSize = 10; blocks = read(bimds); figure montage(blocks,BorderSize=5,BackgroundColor="b") end
Create Training, Test, and Calibration Datastores
Create an ImageDatastore object that contains the training tiles.
dsTrainTiles = imageDatastore(normalBlocksDir,IncludeSubfolders=true,FileExtensions=".png",LabelSource="foldernames");
Add one-hot encoded labels to the training data using the transform function. Specify the transform function using the addLabelData helper function.
dsTrainTiles = transform(dsTrainTiles,@addLabelData,IncludeInfo=true);
Similarly, add one-hot encoded labels to the calibration and test data using the transform function and the addLabelData helper function.
dsCal = transform(dsCal,@addLabelData,IncludeInfo=true); dsTest = transform(dsTest,@addLabelData,IncludeInfo=true);
Define EfficientAD Object Detector Network Architecture
Create the EfficientAD anomaly detector using the efficientADAnomalyDetector object. Specify the UseGlobalAnomalyMap argument as false to turn off the autoencoder and use only the student-teacher model. This enables the model to generalize learned features from training on image tiles to the full-resolution image at inference time.
untrainedDetector = efficientADAnomalyDetector(Network="pdn-small",UseGlobalAnomalyMap=false);Specify Training Options and Train Detector
To train the detector, set the doTraining variable to true.
Train the EfficientAD anomaly detector by using the trainEfficientADAnomalyDetector function. Specify the MapNormalizationDataRatio argument as 0.2 to compute percentile normalization statistics for the student model for 20% of the training data. For the rest of the training data, the autoencoder is turned off and the detector does not update its weights or perform normalization.
Specify network training options using the trainingOptions (Deep Learning Toolbox) function. Specify the Metrics argument as the aucMetric (Deep Learning Toolbox) function handle to monitor the AUC metric, or the area under receiver operating characteristic (ROC) curve, during training. Specify the miniBatchSize name-value argument as 4. If your computing device memory is limited, decrease the mini-batch size to 1 to prevent out-of-memory errors.
Train on one or more GPUs, if they are available. Using a GPU requires a Parallel Computing Toolbox license and a CUDA® enabled NVIDIA® GPU. For more information, see GPU Computing Requirements (Parallel Computing Toolbox). If you have a Parallel Computing Toolbox license, specify the PreprocessingEnvironment argument as "parallel" to accelerate training speed.
doTraining =false; if doTraining maximumEpochs=20; options = trainingOptions("adam", ... InitialLearnRate=1e-4, ... L2Regularization=1e-5, ... LearnRateSchedule="piecewise", ... LearnRateDropPeriod=floor(0.95*maximumEpochs), ... LearnRateDropFactor=0.1, ... MaxEpochs=maximumEpochs, ... VerboseFrequency=2, ... MiniBatchSize=4, ... Shuffle="every-epoch", ... ValidationData=dsCal, ... ValidationPatience=Inf, ... OutputNetwork="best-validation", ... Metrics=aucMetric(Name="auc"), ... ObjectiveMetricName="auc", ... ResetInputNormalization=true,... Plots="training-progress", ... PreprocessingEnvironment="background"); detector = trainEfficientADAnomalyDetector(dsTrainTiles,untrainedDetector,options,MapNormalizationDataRatio=0.2); modelDateTime = string(datetime("now",Format="yyyy-MM-dd-HH-mm-ss")); save(string(tempdir) + filesep + "trainedGumDefectDetectorEfficientAD_" + modelDateTime + ".mat", ... "detector") end
Set Anomaly Threshold
During semi-supervised anomaly detection, you must choose an anomaly score threshold for separating normal images from anomalous images. Select an anomaly score threshold for the anomaly detector, which classifies images based on whether their scores are above or below the threshold value. This example uses the calibration data set defined in the Partition Data into Calibration and Test Sets section, which contains both normal and anomalous images to select the threshold.
Obtain the maximum anomaly score and ground truth label for each image in the calibration set by using the predict object function. Specify the MiniBatchSize argument value as 4, or another small number. Note that the value of this argument must be the same as the mini-batch size used for training in the Specify Training Options and Train Detector section. Because the predict function evaluates the calibration images at full-size resolution, you must specify a small mini-batch size to prevent out-of-memory errors.
scoresCal = predict(detector,dsCal,MiniBatchSize=4);
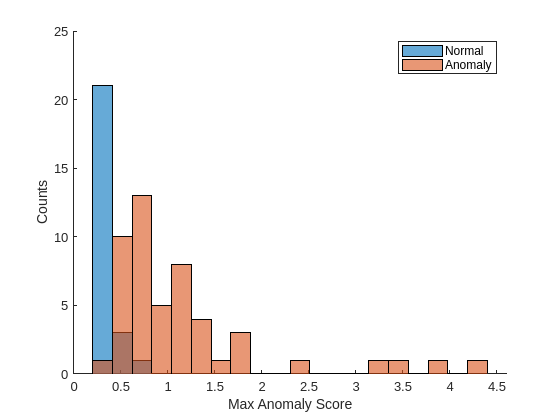
labelsCal = dsCal.UnderlyingDatastores{1}.Labels~="good";Plot a histogram of the maximum anomaly scores for the normal and anomalous classes. The distributions are well-separated by the model-predicted anomaly score.
numBins = 20; [~,edges] = histcounts(scoresCal,numBins); figure hold on hNormal = histogram(scoresCal(labelsCal==0),edges); hAnomaly = histogram(scoresCal(labelsCal==1),edges); hold off legend([hNormal,hAnomaly],"Normal","Anomaly") xlabel("Max Anomaly Score") ylabel("Counts")
Calculate the optimal anomaly threshold by using the anomalyThreshold function. Specify the first two input arguments as the ground truth labels, labels, and predicted anomaly scores scores for the calibration data set. Specify the third input argument as true because true positive anomaly images have a labels value of true. The anomalyThreshold function returns the optimal threshold value as a scalar and the receiver operating characteristic (ROC) curve for the detector as an rocmetrics (Deep Learning Toolbox) object.
[thresh,roc] = anomalyThreshold(labelsCal,scoresCal,true);
Set the Threshold property of the anomaly detector to the optimal threshold value.
detector.Threshold = thresh;
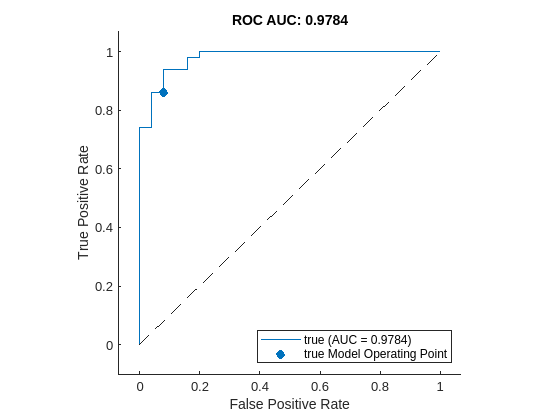
Plot the ROC curve by using the plot (Deep Learning Toolbox) object function of rocmetrics. The ROC curve illustrates the performance of the classifier for a range of possible threshold values. Each point on the ROC curve represents the false positive rate (x-coordinate) and true positive rate (y-coordinate) when you classify the calibration set images using a different threshold value. The solid blue line represents the ROC curve. The area under the ROC curve (AUC) metric indicates classifier performance, with a perfect classifier corresponding to the maximum ROC AUC 1.0.
plot(roc)
title("ROC AUC: " + roc.AUC)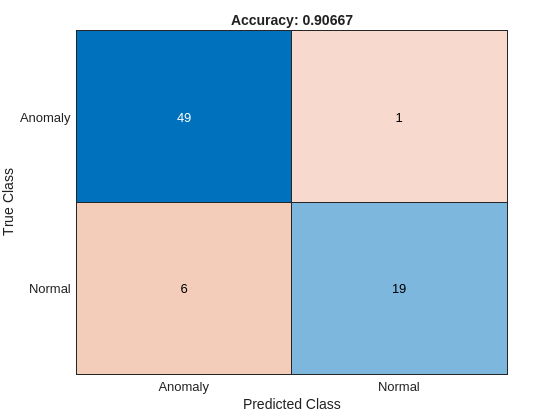
Evaluate Classification Model
Classify each image in the test set as either normal or anomalous by using the classify object function. Specify the MiniBatchSize argument as 1. Because the classify function evaluates the test images at full-size resolution, you must specify a small mini-batch size to prevent out-of-memory errors.
testSetPredictedLabels = classify(detector,dsTest,MiniBatchSize=1); testSetPredictedLabels = testSetPredictedLabels';
Get the ground truth labels of each test image.
testSetGTLabels = dsTest.UnderlyingDatastores{1}.Labels ~= "good";Evaluate the anomaly detector by calculating performance metrics using the evaluateAnomalyDetection function. The function calculates several metrics that evaluate the accuracy, precision, sensitivity, and specificity of the detector for the test data set.
metrics = evaluateAnomalyDetection(testSetPredictedLabels,testSetGTLabels,1);
Evaluating anomaly detection results
------------------------------------
* Finalizing... Done.
* Data set metrics:
GlobalAccuracy MeanAccuracy Precision Recall Specificity F1Score FalsePositiveRate FalseNegativeRate
______________ ____________ _________ ______ ___________ _______ _________________ _________________
0.90667 0.87 0.89091 0.98 0.76 0.93333 0.24 0.02
The ConfusionMatrix property of metrics contains the confusion matrix for the test set. Extract the confusion matrix and display a confusion plot. The classification model in this example is very accurate and predicts a small percentage of false positives and false negatives.
M = metrics.ConfusionMatrix{:,:};
confusionchart(M,["Normal","Anomaly"])
acc = sum(diag(M))/sum(M,"all");
title("Accuracy: " + acc)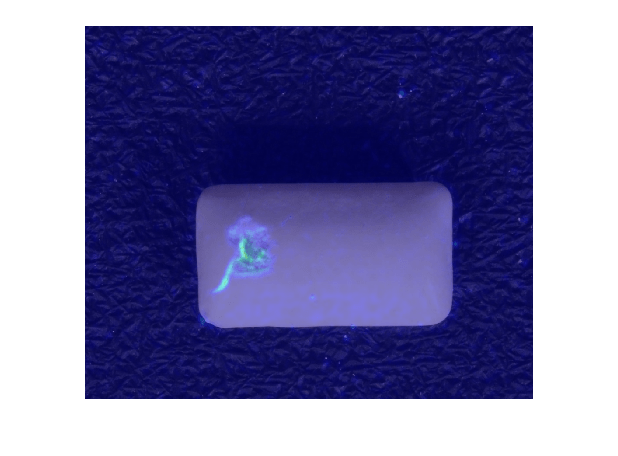
Explain Classification Decisions
You can use the anomaly heatmap that the anomaly detector predicts to explain why the detector classifies an image as normal or anomalous. This approach is useful for identifying patterns in false negatives and false positives. You can use these patterns to identify strategies for increasing class balancing of the training data or improving the network performance.
Normalize Anomaly Heat Map Display Range
To compare anomaly scores observed across the entire calibration set, including normal and anomalous images, normalize the anomaly score map display range. By using the same display range across images, you can compare images more easily than if you scale each image to its own minimum and maximum.
Define a normalized display range, displayRange, that reflects the range of anomaly scores observed across the entire calibration set, including normal and anomalous images.
minMapVal = inf; maxMapVal = -inf; reset(dsCal) while hasdata(dsCal) data = read(dsCal); img = data{1}; map = anomalyMap(detector,img); minMapVal = min(min(map,[],"all"),minMapVal); maxMapVal = max(max(map,[],"all"),maxMapVal); end displayRange = [minMapVal 0.7*maxMapVal];
View Heatmap of Anomalous Image
Select an image of a correctly classified anomaly. Display the image, with the heatmap overlaid by using the anomalyMapOverlay function.
testSetAnomalyLabels = testSetGTLabels;
testSetOutputLabels = testSetPredictedLabels;
idxTruePositive = find(testSetAnomalyLabels & testSetOutputLabels);
dsExample = subset(dsTest,idxTruePositive);
data = read(dsExample);
img = data{1};
map = anomalyMap(detector,img);
imshow(anomalyMapOverlay(img,map,MapRange=displayRange,Blend="equal"))
View Heatmap of Normal Image
Select and display an image of a correctly classified normal image, with the heatmap overlaid by using the anomalyMapOverlay function.
idxTrueNegative = find(~(testSetAnomalyLabels | testSetOutputLabels));
dsExample = subset(dsTest,idxTrueNegative);
data = read(dsExample);
img = data{1};
map = anomalyMap(detector,img);
imshow(anomalyMapOverlay(img,map,MapRange=displayRange,Blend="equal"))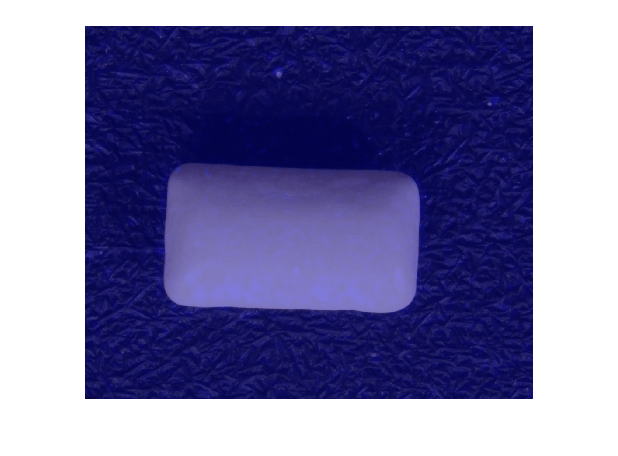
View Heatmap of False Positive Image
False positives are images without defects, but that the network misclassifies as anomalous. Use the explanation from the EfficientAD model [1] to gain insight into the misclassifications.
Select a false positive image, and display it, with the heatmap overlaid by using the anomalyMapOverlay function. For this test image, anomalous scores are localized to image areas with uneven lightning, so you might consider adjusting the image contrast during preprocessing, increasing the number of images used for training, or choosing a different threshold at the calibration step.
idxFalsePositive = find(~(testSetAnomalyLabels) & testSetOutputLabels); if ~isempty(idxFalsePositive) dsExample = subset(dsTest,idxFalsePositive); data = read(dsExample); img = data{1}; map = anomalyMap(detector,img); figure imshow(anomalyMapOverlay(img,map,MapRange=displayRange,Blend="equal")) end

View Heatmap of False Negative Image
False negatives are images with defects, but that the network misclassifies as normal. Use the explanation from the EfficientAD model [1] to gain insights into the misclassifications.
Select a false negative image, and display it, with the heatmap overlaid by using the anomalyMapOverlay function. To decrease false negative results, consider adjusting the anomaly threshold or CompressionRatio of the detector.
idxFalseNegative = find(testSetAnomalyLabels & (~testSetOutputLabels)); if ~isempty(idxFalseNegative) dsExample = subset(dsTest,idxFalseNegative); data = read(dsExample); img = data{1}; map = anomalyMap(detector,img); figure imshow(anomalyMapOverlay(img,map,MapRange=displayRange,Blend="equal")) end

Supporting Functions
addLabelData
The addLabelData helper function creates a one-hot encoded representation of label information in data.
function [data,info] = addLabelData(data,info) classNames = [info.Label]; onehotencoding = classNames ~= categorical("good"); data = {data,onehotencoding}; end
maskObjectsInNormalImages
The maskObjectsInNormalImages helper function creates a logical mask from a blocked image, in which true (or 1) pixels represent ROIs with meaningful normal data.
function bout = maskObjectsInNormalImages(bim,T) im = im2gray(bim.Data); % Threshold image using global threshold BW = imbinarize(im,T); % Morphologically open mask with square-shaped % structuring element width = 3; se = strel("square",width); BW = imopen(BW,se); bout = BW; end
References
[1] Batzner, Kilian, Lars Heckler, and Rebecca König. “EfficientAD: Accurate Visual Anomaly Detection at Millisecond-Level Latencies.” In 2024 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), 127–37. Waikoloa, HI, USA: IEEE, 2024. https://doi.org/10.1109/WACV57701.2024.00020.
[2] Zou, Yang, Jongheon Jeong, Latha Pemula, Dongqing Zhang, and Onkar Dabeer. “SPot-the-Difference Self-Supervised Pre-Training for Anomaly Detection and Segmentation.” arXiv, July 28, 2022. https://doi.org/10.48550/arXiv.2207.14315.
See Also
trainEfficientADAnomalyDetector | efficientADAnomalyDetector | predict | anomalyMap | evaluateAnomalyDetection | trainingOptions (Deep Learning Toolbox) | anomalyThreshold