forward
Compute deep learning network output for training
Syntax
Description
Some deep learning layers behave differently during training and inference (prediction). For example, during training, dropout layers randomly set input elements to zero to help prevent overfitting, but during inference, dropout layers do not change the input.
To compute network outputs for training, use the forward function. To
compute network outputs for inference, use the predict function.
[
also returns a cell array of activations of the pruning layers. This syntax is applicable
only if Y1,...,YN,state,pruningActivations] = forward(X1,...,XM)net is a TaylorPrunableNetwork object.
To prune a deep neural network, you require the Deep Learning Toolbox™ Model Compression Library support package. This support package is a free add-on that you can download using the Add-On Explorer. Alternatively, see Deep Learning Toolbox Model Compression Library.
___ = forward(___,
specifies additional options using one or more name-value arguments.Name=Value)
Examples
This example shows how to train a network that classifies handwritten digits with a custom learning rate schedule.
You can train most types of neural networks using the trainnet and trainingOptions functions. If the trainingOptions function does not provide the options you need (for example, a custom solver), then you can define your own custom training loop using dlarray and dlnetwork objects for automatic differentiation. For an example showing how to retrain a pretrained deep learning network using the trainnet function, see Retrain Neural Network to Classify New Images.
Training a deep neural network is an optimization task. By considering a neural network as a function , where is the network input, and is the set of learnable parameters, you can optimize so that it minimizes some loss value based on the training data. For example, optimize the learnable parameters such that for a given inputs with a corresponding targets , they minimize the error between the predictions and .
The loss function used depends on the type of task. For example:
For classification tasks, you can minimize the cross entropy error between the predictions and targets.
For regression tasks, you can minimize the mean squared error between the predictions and targets.
You can optimize the objective using gradient descent: minimize the loss by iteratively updating the learnable parameters by taking steps towards the minimum using the gradients of the loss with respect to the learnable parameters. Gradient descent algorithms typically update the learnable parameters by using a variant of an update step of the form , where is the iteration number, is the learning rate, and denotes the gradients (the derivatives of the loss with respect to the learnable parameters).
This example trains a network to classify handwritten digits with the stochastic gradient descent algorithm (without momentum).
Load Training Data
Load the digits data as an image datastore using the imageDatastore function and specify the folder containing the image data.
unzip("DigitsData.zip") imds = imageDatastore("DigitsData", ... IncludeSubfolders=true, ... LabelSource="foldernames");
Partition the data into training and test sets. Set aside 10% of the data for testing using the splitEachLabel function.
[imdsTrain,imdsTest] = splitEachLabel(imds,0.9,"randomize");The network used in this example requires input images of size 28-by-28-by-1. To automatically resize the training images, use an augmented image datastore. Specify additional augmentation operations to perform on the training images: randomly translate the images up to 5 pixels in the horizontal and vertical axes. Data augmentation helps prevent the network from overfitting and memorizing the exact details of the training images.
inputSize = [28 28 1]; pixelRange = [-5 5]; imageAugmenter = imageDataAugmenter( ... RandXTranslation=pixelRange, ... RandYTranslation=pixelRange); augimdsTrain = augmentedImageDatastore(inputSize(1:2),imdsTrain,DataAugmentation=imageAugmenter);
To automatically resize the test images without performing further data augmentation, use an augmented image datastore without specifying any additional preprocessing operations.
augimdsTest = augmentedImageDatastore(inputSize(1:2),imdsTest);
Determine the number of classes in the training data.
classes = categories(imdsTrain.Labels); numClasses = numel(classes);
Define Network
Define the network for image classification.
For image input, specify an image input layer with input size matching the training data.
Do not normalize the image input, set the
Normalizationoption of the input layer to"none".Specify three convolution-batchnorm-ReLU blocks.
Pad the input to the convolution layers such that the output has the same size by setting the
Paddingoption to"same".For the first convolution layer specify 20 filters of size 5. For the remaining convolution layers specify 20 filters of size 3.
For classification, specify a fully connected layer with size matching the number of classes
To map the output to probabilities, include a softmax layer.
When training a network using a custom training loop, do not include an output layer.
layers = [
imageInputLayer(inputSize,Normalization="none")
convolution2dLayer(5,20,Padding="same")
batchNormalizationLayer
reluLayer
convolution2dLayer(3,20,Padding="same")
batchNormalizationLayer
reluLayer
convolution2dLayer(3,20,Padding="same")
batchNormalizationLayer
reluLayer
fullyConnectedLayer(numClasses)
softmaxLayer];Create a dlnetwork object from the layer array.
net = dlnetwork(layers)
net =
dlnetwork with properties:
Layers: [12×1 nnet.cnn.layer.Layer]
Connections: [11×2 table]
Learnables: [14×3 table]
State: [6×3 table]
InputNames: {'imageinput'}
OutputNames: {'softmax'}
Initialized: 1
View summary with summary.
Define Model Loss Function
Training a deep neural network is an optimization task. By considering a neural network as a function , where is the network input, and is the set of learnable parameters, you can optimize so that it minimizes some loss value based on the training data. For example, optimize the learnable parameters such that for a given inputs with a corresponding targets , they minimize the error between the predictions and .
Define the modelLoss function. The modelLoss function takes a dlnetwork object net, a mini-batch of input data X with corresponding targets T and returns the loss, the gradients of the loss with respect to the learnable parameters in net, and the network state. To compute the gradients automatically, use the dlgradient function.
function [loss,gradients,state] = modelLoss(net,X,T) % Forward data through network. [Y,state] = forward(net,X); % Calculate cross-entropy loss. loss = crossentropy(Y,T); % Calculate gradients of loss with respect to learnable parameters. gradients = dlgradient(loss,net.Learnables); end
Define SGD Function
Create the function sgdStep that takes the parameters and the gradients of the loss with respect to the parameters, and returns the updated parameters using the stochastic gradient descent algorithm, expressed as , where is the iteration number, is the learning rate, and denotes the gradients (the derivatives of the loss with respect to the learnable parameters).
function parameters = sgdStep(parameters,gradients,learnRate) parameters = parameters - learnRate .* gradients; end
Defining a custom update function is not a necessary step for custom training loops. Alternatively, you can use built in update functions like sgdmupdate, adamupdate, and rmspropupdate.
Specify Training Options
Train for fifteen epochs with a mini-batch size of 128 and a learning rate of 0.01.
numEpochs = 15; miniBatchSize = 128; learnRate = 0.01;
Train Model
Create a minibatchqueue object that processes and manages mini-batches of images during training. For each mini-batch:
Use the custom mini-batch preprocessing function
preprocessMiniBatch(defined at the end of this example) to convert the targets to one-hot encoded vectors.Format the image data with the dimension labels
"SSCB"(spatial, spatial, channel, batch). By default, theminibatchqueueobject converts the data todlarrayobjects with underlying typesingle. Do not format the targets.Discard partial mini-batches.
Train on a GPU if one is available. By default, the
minibatchqueueobject converts each output to agpuArrayif a GPU is available. Using a GPU requires Parallel Computing Toolbox™ and a supported GPU device. For information on supported devices, see GPU Computing Requirements (Parallel Computing Toolbox).
mbq = minibatchqueue(augimdsTrain,... MiniBatchSize=miniBatchSize,... MiniBatchFcn=@preprocessMiniBatch,... MiniBatchFormat=["SSCB" ""], ... PartialMiniBatch="discard");
Calculate the total number of iterations for the training progress monitor.
numObservationsTrain = numel(imdsTrain.Files); numIterationsPerEpoch = floor(numObservationsTrain / miniBatchSize); numIterations = numEpochs * numIterationsPerEpoch;
Initialize the TrainingProgressMonitor object. Because the timer starts when you create the monitor object, make sure that you create the object close to the training loop.
monitor = trainingProgressMonitor( ... Metrics="Loss", ... Info="Epoch", ... XLabel="Iteration");
Train the network using a custom training loop. For each epoch, shuffle the data and loop over mini-batches of data. For each mini-batch:
Evaluate the model loss, gradients, and state using the
dlfevalandmodelLossfunctions and update the network state.Update the network parameters using the
dlupdatefunction with the custom update function.Update the loss and epoch values in the training progress monitor.
Stop if the Stop property of the monitor is true. The Stop property value of the
TrainingProgressMonitorobject changes to true when you click the Stop button.
epoch = 0; iteration = 0; % Loop over epochs. while epoch < numEpochs && ~monitor.Stop epoch = epoch + 1; % Shuffle data. shuffle(mbq); % Loop over mini-batches. while hasdata(mbq) && ~monitor.Stop iteration = iteration + 1; % Read mini-batch of data. [X,T] = next(mbq); % Evaluate the model gradients, state, and loss using dlfeval and the % modelLoss function and update the network state. [loss,gradients,state] = dlfeval(@modelLoss,net,X,T); net.State = state; % Update the network parameters using SGD. updateFcn = @(parameters,gradients) sgdStep(parameters,gradients,learnRate); net = dlupdate(updateFcn,net,gradients); % Update the training progress monitor. recordMetrics(monitor,iteration,Loss=loss); updateInfo(monitor,Epoch=epoch); monitor.Progress = 100 * iteration/numIterations; end end

Test Model
Test the neural network using the testnet function. For single-label classification, evaluate the accuracy. The accuracy is the percentage of correct predictions. By default, the testnet function uses a GPU if one is available. To select the execution environment manually, use the ExecutionEnvironment argument of the testnet function.
accuracy = testnet(net,augimdsTest,"accuracy")accuracy = 96.3000
Visualize the predictions in a confusion chart. Make predictions using the minibatchpredict function, and convert the classification scores to labels using the scores2label function. By default, the minibatchpredict function uses a GPU if one is available. To select the execution environment manually, use the ExecutionEnvironment argument of the minibatchpredict function.
scores = minibatchpredict(net,augimdsTest); YTest = scores2label(scores,classes);
Visualize the predictions in a confusion chart.
TTest = imdsTest.Labels; figure confusionchart(TTest,YTest)
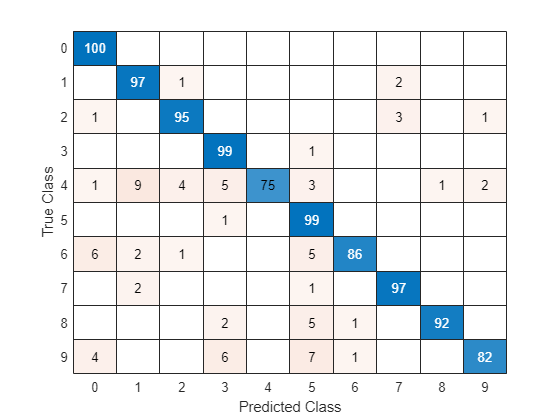
Large values on the diagonal indicate accurate predictions for the corresponding class. Large values on the off-diagonal indicate strong confusion between the corresponding classes.
Supporting Functions
Mini Batch Preprocessing Function
The preprocessMiniBatch function preprocesses a mini-batch of predictors and labels using the following steps:
Preprocess the images using the
preprocessMiniBatchPredictorsfunction.Extract the label data from the incoming cell array and concatenate into a categorical array along the second dimension.
One-hot encode the categorical labels into numeric arrays. Encoding into the first dimension produces an encoded array that matches the shape of the network output.
function [X,T] = preprocessMiniBatch(dataX,dataT) % Preprocess predictors. X = preprocessMiniBatchPredictors(dataX); % Extract label data from cell and concatenate. T = cat(2,dataT{:}); % One-hot encode labels. T = onehotencode(T,1); end
Mini-Batch Predictors Preprocessing Function
The preprocessMiniBatchPredictors function preprocesses a mini-batch of predictors by extracting the image data from the input cell array and concatenate into a numeric array. For grayscale input, concatenating over the fourth dimension adds a third dimension to each image, to use as a singleton channel dimension.
function X = preprocessMiniBatchPredictors(dataX) % Concatenate. X = cat(4,dataX{:}); end
Input Arguments
Neural network, specified as one of these values:
dlnetworkobject — Neural network for custom training loop.TaylorPrunableNetworkobject — Neural network for custom pruning loop.
To prune a deep neural network, you require the Deep Learning Toolbox Model Compression Library support package. This support package is a free add-on that you can download using the Add-On Explorer. Alternatively, see Deep Learning Toolbox Model Compression Library.
Input data, each specified as one of these values:
Formatted
dlarrayobjectUnformatted
dlarrayobject (since R2025a)Numeric array (since R2025a)
The input Xi corresponds to the network input
net.InputNames(i).
To train a neural network using a custom training loop using automatic
differentiation, the input data must be formatted or unformatted
dlarray objects.
Tip
Neural networks expect input data with a specific layout. For example, vector-sequence classification networks typically expect vector-sequence representations to be t-by-c arrays, where t and c are the number of time steps and channels of sequences, respectively. Neural networks typically have an input layer that specifies the expected layout of the data.
Most datastores and functions output data in the layout that the network expects. If
your data is in a different layout to what the network expects, then indicate that your
data has a different layout by using the InputDataFormats option or
by specifying input data as a formatted dlarray object. It is usually
easiest to adjust the InputDataFormats training option than to
preprocess the input data.
For more information, see Deep Learning Data Formats.
To create a neural network that receives unformatted
data, use an inputLayer object
and do not specify a format. To input unformatted data into a network directly, do not
specify the InputDataFormats argument. (since R2025a)
Before R2025a: For neural networks that do not have input layers, you
must specify a format using the InputDataFormats argument or use
formatted dlarray objects as input.
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: forward(net,X,Outputs=["relu1" "relu2"]) specifies to
extract outputs from the layers with the names "relu1" and
"relu2".
Neural network outputs, specified as a string array or a cell array of character vectors of layer names or layer output paths. Specify the output using one of these forms:
"layerName", wherelayerNameis the name of a layer with a single output."layerName/outputName", wherelayerNameis the name of a layer andoutputNameis the name of the layer output. Use this option for layers with multiple outputs.
To use the outputs of layers inside a networkLayer
object, first expand the nested network using the expandLayers function.
If you do not specify the layers to extract outputs from, then, by default, the
software uses the outputs specified by net.Outputs.
Performance optimization, specified as one of these values:
"auto"– Automatically apply a number of optimizations suitable for the input network and hardware resources."none"– Disable all optimizations.
Using the "auto" acceleration option can offer performance benefits,
but at the expense of an increased initial run time. Subsequent calls with compatible
parameters are faster. Use performance optimization when you plan to call the function
multiple times using different input data with the same size and shape.
Since R2025a
Description of the input data dimensions, specified as a string array, character vector, or cell array of character vectors.
If InputDataFormats is "auto", then the software uses
the formats expected by the network input. Otherwise, the software uses the specified
formats for the corresponding network input.
A data format is a string of characters, where each character describes the type of the corresponding data dimension.
The characters are:
"S"— Spatial"C"— Channel"B"— Batch"T"— Time"U"— Unspecified
For example, consider an array that represents a batch of sequences where the first,
second, and third dimensions correspond to channels, observations, and time steps,
respectively. You can describe the data as having the format "CBT"
(channel, batch, time).
You can specify multiple dimensions labeled "S" or "U".
You can use the labels "C", "B", and
"T" once each, at most. The software ignores singleton trailing
"U" dimensions after the second dimension.
For a neural networks with multiple inputs net, specify an array of
input data formats, where InputDataFormats(i) corresponds to the
input net.InputNames(i).
For more information, see Deep Learning Data Formats.
To create a neural network that receives unformatted
data, use an inputLayer object
and do not specify a format. To input unformatted data into a network directly, do not
specify the InputDataFormats argument. (since R2025a)
Before R2025a: For neural networks that do not have input layers, you
must specify a format using the InputDataFormats argument or use
formatted dlarray objects as input.
Data Types: char | string | cell
Since R2025a
Description of the output data dimensions, specified as one of these values:
"auto"— If the output data has the same number of dimensions as the input data, then theforwardfunction uses the format specified byInputDataFormats. If the output data has a different number of dimensions than the input data, then theforwardfunction automatically permutes the dimensions of the output data so that they are consistent with the network input layers or theInputDataFormatsvalue.String, character vector, or cell array of character vectors — The
forwardfunction uses the specified data formats.
A data format is a string of characters, where each character describes the type of the corresponding data dimension.
The characters are:
"S"— Spatial"C"— Channel"B"— Batch"T"— Time"U"— Unspecified
For example, consider an array that represents a batch of sequences where the first,
second, and third dimensions correspond to channels, observations, and time steps,
respectively. You can describe the data as having the format "CBT"
(channel, batch, time).
You can specify multiple dimensions labeled "S" or "U".
You can use the labels "C", "B", and
"T" once each, at most. The software ignores singleton trailing
"U" dimensions after the second dimension.
For more information, see Deep Learning Data Formats.
Data Types: char | string | cell
Output Arguments
Output data of network with multiple outputs, returned as a one of these values:
Formatted
dlarrayobjectUnformatted
dlarrayobject (since R2025a)Numeric array (since R2025a)
The data type matches the data type of the input data.
The order of the outputs Y1, …, YN match the
order of the outputs specified by the Outputs argument.
For a classification neural network, the elements of the output correspond to the scores for
each class. The order of the scores matches the order of the categories in the training
data. For example, if you train the neural network using the categorical labels
TTrain, then the order of the scores matches the order of the
categories given by categories(TTrain).
Updated network state, returned as a table.
The network state is a table with three columns:
Layer– Layer name, specified as a string scalar.Parameter– State parameter name, specified as a string scalar.Value– Value of state parameter, specified as adlarrayobject.
Layer states retain information calculated during the layer operation for use in subsequent forward passes of the layer. For example, LSTM layers contain cell states and hidden states, and batch normalization layers calculate running statistics.
For recurrent layers, such as LSTM layers, with the HasStateInputs
property set to 1 (true), the state table does
not contain entries for the states of the layer.
Cell array of activations of the pruning layers, if the input network is a TaylorPrunableNetwork object.
Extended Capabilities
The forward function
supports GPU array input with these usage notes and limitations:
This function runs on the GPU if you meet at least one of these conditions:
Any of the values of the network learnable parameters inside
net.Learnables.Valuearedlarrayobjects with underlying data of typegpuArray.The input argument
Xis adlarraywith underlying data of typegpuArray.
For more information, see Run MATLAB Functions on a GPU (Parallel Computing Toolbox).
Version History
Introduced in R2019bOptionally, specify the input and output data formats using the
InputDataFormats and OutputDataFormats options,
respectively. If you specify unformatted data as input to the neural network and do not
specify the InputDataFormats argument, then the function passes the
unformatted data to the network directly.
To create a neural network that receives unformatted data, use an inputLayer object
and do not specify a format.
For dlnetwork objects, the state output argument returned by the forward function is
a table containing the state parameter names and values for each layer in the network.
Starting in R2021a, the state values are dlarray objects.
This change enables better support when using AcceleratedFunction
objects. To accelerate deep learning functions that have frequently changing input values,
for example, an input containing the network state, the frequently changing values must be
specified as dlarray objects.
In previous versions, the state values are numeric arrays.
In most cases, you will not need to update your code. If you have code that requires the
state values to be numeric arrays, then to reproduce the previous behavior, extract the data
from the state values manually using the extractdata
function with the dlupdate
function.
state = dlupdate(@extractdata,net.State);
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)