自组织映射神经网络的聚类
自组织特征映射 (SOFM) 学习根据输入向量在输入空间中的分组方式对输入向量进行分类。它们与竞争层的不同之处在于,自组织映射中的相邻神经元会学习识别输入空间的相邻部分。因此,自组织映射会同时学习训练时所基于的输入向量的分布(如竞争层所做的一样)和拓扑。
SOFM 的层中的神经元最初根据拓扑函数排列在物理位置中。函数 gridtop、hextop 或 randtop 可以将神经元排列成网格、六边形或随机拓扑。神经元之间的距离是使用距离函数根据其位置进行计算的。有四个距离函数:dist、boxdist、linkdist 和 mandist。连接距离是最常见的。这些拓扑和距离函数在拓扑(gridtop、hextop、randtop)和距离函数(dist、linkdist、mandist、boxdist)中进行了介绍。
此处,自组织特征映射网络使用与竞争层相同的过程来识别获胜神经元 i*。然而,不是仅更新获胜神经元,而是使用 Kohonen 规则更新获胜神经元的特定邻域 Ni* (d) 内的所有神经元。具体来说,按如下方式调整所有这样的神经元 i ∊ Ni* (d):
或
此处的邻域 Ni* (d) 包含位于获胜神经元 i 的半径 d 范围内的所有神经元的索引*。
因此,当提交向量 p 时,获胜神经元和其近邻的权重向 p 移动。因此,多次提交输入后,邻近神经元就学习了彼此相似的向量。
SOFM 训练的另一个版本称为批量算法,它会在将整个数据集提交给网络后再更新权重。然后,该算法为每个输入向量确定一个获胜神经元。随后将每个权重向量移至特定位置,即它作为获胜神经元或位于获胜神经元的临域时的所有对应输入向量的平均位置。
为了说明邻域的概念,请参考下图。左图显示神经元 13 周围半径 d = 1 的二维邻域。右图显示半径 d = 2 的邻域。
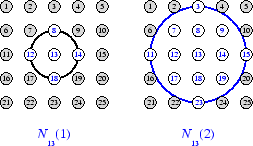
这些邻域可以写为 N13(1) = {8, 12, 13, 14, 18} 和
N13(2) = {3, 7, 8, 9, 11, 12, 13, 14, 15, 17, 18, 19, 23}。
SOFM 中的神经元不必以二维模式排列。您可以使用一维排列,也可以使用三维或多维排列。对于一维 SOFM,一个神经元在半径为 1 的范围内只有两个邻点(如果该神经元位于行的末端,则只有一个邻点)。您也可以用不同方式定义距离,例如,使用神经元和邻域的矩形和六边形排列。网络的性能对邻域的确切形状并不敏感。
拓扑(gridtop、hextop、randtop)
您可以使用函数 gridtop、hextop 和 randtop 为原始神经元位置指定不同拓扑。
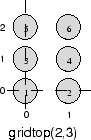
gridtop 拓扑以矩形网格中的神经元开始,类似于上图所示。例如,假设您需要一个由 6 个神经元组成的 2×3 数组。您可以使用以下代码来实现它:
pos = gridtop([2, 3])
pos =
0 1 0 1 0 1
0 0 1 1 2 2
此处,神经元 1 具有位置 (0,0),神经元 2 具有位置 (1,0),神经元 3 具有位置 (0,1) 等。
请注意,如果您要求维度大小颠倒的 gridtop,您会得到稍微不同的排列:
pos = gridtop([3, 2])
pos =
0 1 2 0 1 2
0 0 0 1 1 1
您可以使用以下代码在 gridtop 拓扑中创建一组 8×10 神经元:
pos = gridtop([8 10]); plotsom(pos)

如图所示,gridtop 拓扑中的神经元确实位于一个网格上。
hextop 函数创建一组相似的神经元,但它们采用六边形模式。生成的 2×3 模式的 hextop 神经元如下:
pos = hextop([2, 3])
pos =
0 1.0000 0.5000 1.5000 0 1.0000
0 0 0.8660 0.8660 1.7321 1.7321
请注意,hextop 是使用 selforgmap 生成的 SOM 网络的默认模式。
您可以使用以下代码以 hextop 拓扑创建并绘制一组 8×10 神经元:
pos = hextop([8 10]); plotsom(pos)

注意六边形排列的神经元的位置。
最后,randtop 函数以 N 维随机模式创建神经元。以下代码生成一个随机神经元模式。
pos = randtop([2, 3])
pos =
0 0.7620 0.6268 1.4218 0.0663 0.7862
0.0925 0 0.4984 0.6007 1.1222 1.4228
您可以使用以下代码以 randtop 拓扑创建并绘制一组 8×10 神经元:
pos = randtop([8 10]); plotsom(pos)
有关示例,请参阅这些拓扑函数的帮助。
距离函数(dist、linkdist、mandist、boxdist)
在此工具箱中,有四种方法可以计算特定神经元到其邻点的距离。每种计算方法都用一个特殊函数来实现。
dist 函数计算从主神经元到其他神经元的欧几里德距离。假设您有三个神经元:
pos2 = [0 1 2; 0 1 2]
pos2 =
0 1 2
0 1 2
您可以使用以下代码找到每个神经元与其他神经元之间的距离:
D2 = dist(pos2)
D2 =
0 1.4142 2.8284
1.4142 0 1.4142
2.8284 1.4142 0
因此,神经元 1 到自身的距离是 0,从神经元 1 到神经元 2 的距离是 1.4142 等。
下图显示二维 (gridtop) 神经元层中的一个主神经元。该主神经元周围有直径越来越大的邻域。直径为 1 的邻域包含该主神经元及其紧挨的邻近神经元。直径为 2 的邻域包含直径为 1 的神经元及其紧挨的邻近神经元。
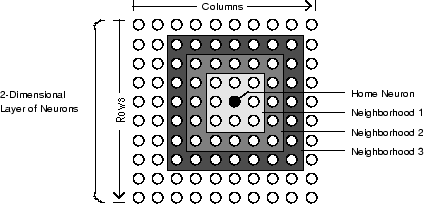
对于 dist 函数,S 神经元层映射的所有邻域都由距离的 S×S 矩阵表示。上面显示的特定距离(在紧挨的邻域中为 1,在邻域 2 中为 2,等等)由函数 boxdist 生成。假设在 gridtop 配置中有六个神经元。
pos = gridtop([2, 3])
pos =
0 1 0 1 0 1
0 0 1 1 2 2
则框距离为
d = boxdist(pos)
d =
0 1 1 1 2 2
1 0 1 1 2 2
1 1 0 1 1 1
1 1 1 0 1 1
2 2 1 1 0 1
2 2 1 1 1 0
从神经元 1 到神经元 2、3 和 4 的距离只有 1,因为它们在紧挨邻域中。神经元 1 到 5 和 6 的距离均为 2。从神经元 3 和 4 到所有其他神经元的距离仅为 1。
到一个神经元的连接距离是到达所关注的神经元所必须经过的连接数或步数。因此,如果您使用 linkdist 计算到同一组神经元的距离,得到的结果如下
dlink =
0 1 1 2 2 3
1 0 2 1 3 2
1 2 0 1 1 2
2 1 1 0 2 1
2 3 1 2 0 1
3 2 2 1 1 0
两个向量 x 和 y 之间的 Manhattan 距离的计算方式如下
D = sum(abs(x-y))
因此,如果您有
W1 = [1 2; 3 4; 5 6]
W1 =
1 2
3 4
5 6
和
P1 = [1;1]
P1 =
1
1
则您得到距离
Z1 = mandist(W1,P1)
Z1 =
1
5
9
用 mandist 计算的距离确实符合上面给出的数学表达式。
架构
此 SOFM 的架构如下所示。
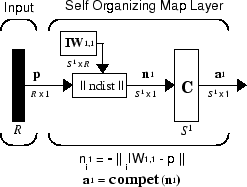
此架构类似于竞争网络的架构,不同之处是此处不使用偏置。对于与 i*(获胜神经元)对应的输出元素 a1i ,竞争传递函数会输出 1。a1 中的所有其他输出元素均为 0。
然而,如上所述,现在靠近获胜神经元的神经元随获胜神经元一起更新。您可以从不同神经元拓扑中进行选择。同样,您可以从各种距离表达式中进行选择,来计算靠近获胜神经元的神经元。
创建自组织映射神经网络 (selforgmap)
您可以使用函数 selforgmap 创建一个新 SOM 网络。该函数定义在两个学习阶段中使用的变量:
排序阶段学习率
排序阶段步骤
调整阶段学习率
调整阶段邻域距离
这些值用于训练和自适应。
请参考以下示例。
假设您要创建一个包含由两个元素组成的输入向量的网络,并且要在一个六边形 2×3 网络中包含六个神经元。获得该网络的代码如下:
net = selforgmap([2 3]);
假设要用于训练的向量是:
P = [.1 .3 1.2 1.1 1.8 1.7 .1 .3 1.2 1.1 1.8 1.7;...
0.2 0.1 0.3 0.1 0.3 0.2 1.8 1.8 1.9 1.9 1.7 1.8];您可以使用以下代码配置网络以输入数据并绘制所有这些数据:
net = configure(net,P); plotsompos(net,P)
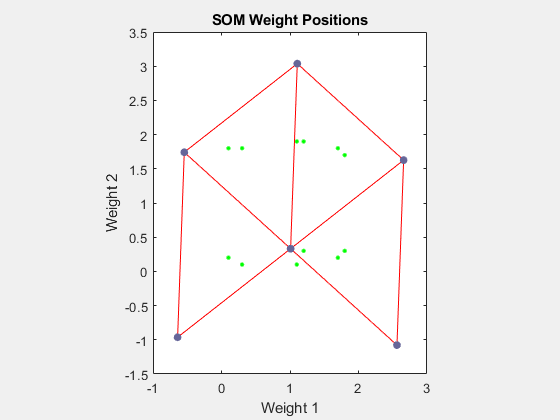
绿点是训练向量。selforgmap 的初始化将初始权重分布到整个输入空间。请注意,它们最初离训练向量有一定距离。
当对网络进行仿真时,计算每个神经元的权重向量和输入向量之间的负距离 (negdist) 以获得加权输入。加权输入也是净输入 (netsum)。净输入相互竞争 (compet),因此只有具有最大正值净输入的神经元才会输出 1。
训练 (learnsomb)
自组织特征映射中的默认学习在批量模式 (trainbu) 下进行。自组织映射的权重学习函数是 learnsomb。
首先,网络为每个输入向量确定获胜神经元。随后将每个权重向量移至特定位置,即它作为获胜神经元或位于获胜神经元的临域时的所有对应输入向量的平均位置。定义邻域大小的距离在训练过程中的两个阶段会更改。
排序阶段
该阶段持续给定的步数。邻域距离从给定的初始距离开始,并减小到调整邻域距离 (1.0)。随着邻域距离在此阶段逐步减小,网络的神经元通常在输入空间中以与它们自己的物理排序相同的拓扑进行排序。
调整阶段
此阶段持续训练或自适应阶段的其余部分。邻域大小已降至 1 以下,因此只有获胜神经元对每个样本进行学习。
现在看看这些网络中常用的一些特定值。
根据 learnsomb 学习参数进行学习,此处显示其默认值。
学习参数 | 默认值 | 目的 |
|---|---|---|
|
| 初始邻域大小 |
|
| 排序阶段步骤 |
邻域大小 NS 在两个阶段中进行更改:排序阶段和调整阶段。
排序阶段持续的步骤与 LP.steps 一样多。在此阶段,算法将 ND 从初始邻域大小 LP.init_neighborhood 向下调整为 1。在此阶段,神经元权重在输入空间中自行排序,且顺序应与相关联的神经元的位置一致。
在调整阶段,ND 小于 1。在此阶段,权重应该在输入空间中相对均匀地分布,同时保持在排序阶段建立的拓扑顺序。
因此,神经元的权重向量最初全部一起以较大步幅向输入空间中出现输入向量的区域前进。然后,当邻域大小减小到 1 时,映射趋于根据提交的输入向量对自身进行拓扑排序。一旦邻域大小为 1,网络应很好地完成排序。训练继续进行,以便给神经元时间来在输入向量中均匀分布。
与竞争层一样,如果输入向量以均匀概率出现在输入空间的一部分中,自组织映射的神经元将按照它们之间近似相等的距离来对自身排序。如果各输入向量在整个输入空间中以不同频率出现,则特征映射层倾向于将神经元分配给与该处输入向量的频率成比例的区域。
因此,特征图在学习对其输入进行分类的同时,也学习其输入的拓扑和分布。
您可以使用以下代码对网络进行 1000 轮训练
net.trainParam.epochs = 1000; net = train(net,P);

plotsompos(net,P)
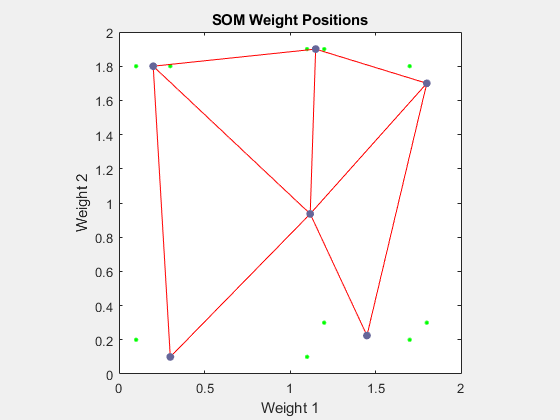
您可以看到神经元已开始向不同训练组移动。需要额外的训练来使神经元更接近不同组。
如前所述,在神经元更新其权重方面,自组织映射不同于传统的竞争学习。特征图不是只更新获胜神经元,而是更新获胜神经元及其邻点的权重。结果是相邻的神经元倾向于具有相似的权重向量,并对相似的输入向量作出响应。
示例
下面简要介绍两个示例。您也可以尝试类似的示例一维自组织映射和二维自组织映射。
一维自组织映射
假设 100 个二元素单位输入向量均匀分布在 0° 和 90° 之间。
angles = 0:0.5*pi/99:0.5*pi;
以下是数据图。
P = [sin(angles); cos(angles)];
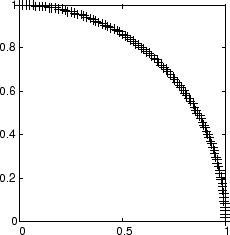
自组织映射定义为一个包含 10 个神经元的一维层。该图将根据上述输入向量进行训练。最初,这些神经元位于图形的中心。
当然,由于所有权重向量都从输入向量空间的中间开始,您现在看到的只是一个圆。
随着训练的开始,权重向量一起向输入向量移动。随着邻域大小的减小,它们也会进行排序。最后,该层会调整其权重,使得每个神经元对输入向量占据的输入空间区域作出强响应。相邻神经元权重向量的位置也反映输入向量的拓扑。
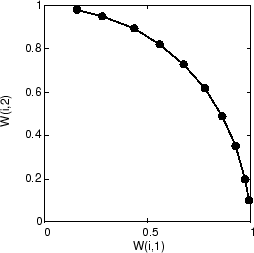
请注意,自组织映射是用随机顺序的输入向量来训练的,因此从相同的初始向量开始并不能保证获得相同的训练结果。
二维自组织映射
此示例说明如何训练二维自组织映射。
首先,用以下代码创建一些随机输入数据:
P = rands(2,1000);
这是以下 1000 个输入向量的图。
![Plot of 1000 random input vectors between [-1 -1] and [1 1].](selforg_demsm2a.gif)
使用一个包含 30 个神经元的 5×6 二维图对这些输入向量进行分类。该二维图由 5×6 神经元组成,根据 Manhattan 距离邻域函数 mandist 计算距离。
然后,通过 5000 个数据提交周期来训练映射,每 20 个周期显示一次。
这是 40 个周期后的自组织映射。
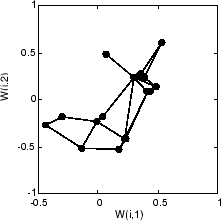
用圆表示的权重向量几乎是随机放置的。然而,即使只经过 40 个数据提交周期,由线连接的相邻神经元也有了紧密靠近的权重向量。
以下是 120 个周期后的图。
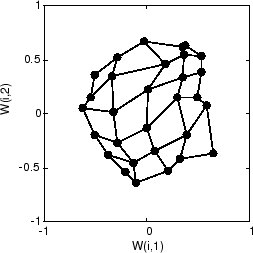
120 个周期后,图开始根据输入空间的拓扑来组织自身,这将限制输入向量。
以下是在 500 个周期后在输入空间中更均匀分布的图。
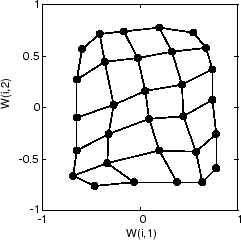
最后,以下是在 5000 个周期后在输入空间中分布相当均匀的图。此外,神经元的间距非常均匀,反映在此问题中输入向量等间距。
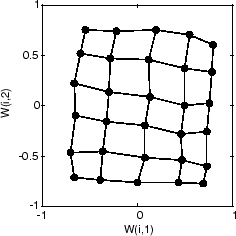
因此,二维自组织映射已学习了其输入空间的拓扑。
必须注意,虽然自组织映射不需要很长时间来组织自身即可使相邻神经元识别相似的输入,但映射最终根据输入向量的分布来排列自身可能需要很长时间。
用批量算法进行训练
批量训练算法通常比增量算法快得多,并且是 SOFM 训练的默认算法。您可以使用以下命令对简单数据集试验该算法:
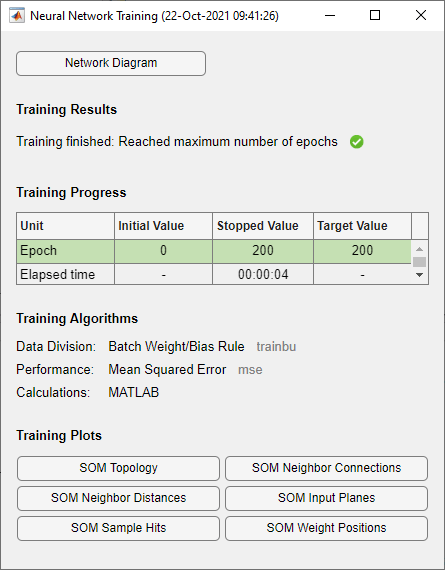
x = simplecluster_dataset net = selforgmap([6 6]); net = train(net,x);
此命令序列创建并训练一个由 36 个神经元组成的 6×6 二维图。在训练过程中,出现以下图窗。
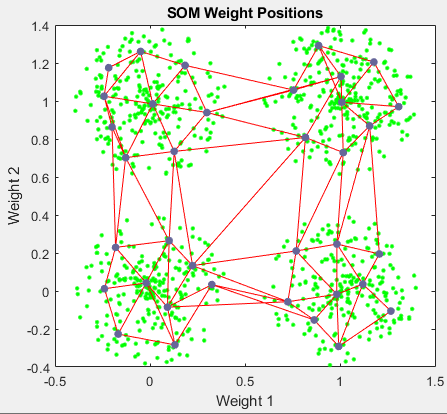
您可以从该窗口访问几个有用的可视化。如果您点击 SOM 权重位置,将出现以下图窗,其中显示数据点和权重向量的位置。如该图窗所示,批量算法仅经过 200 次迭代,图就在输入空间中得到很好的分布。
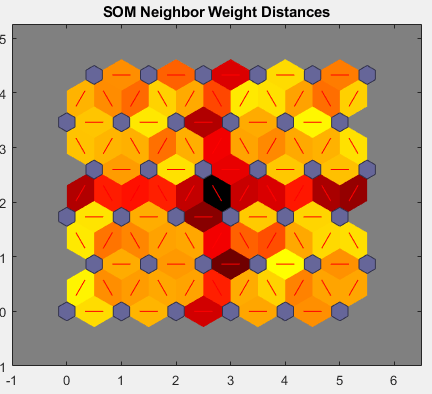
当输入空间是高维空间时,您无法同时可视化所有权重。在本例中,请点击 SOM 邻点距离。以下图窗显示相邻神经元之间的距离。
该图窗使用以下着色方案:
蓝色六边形表示神经元。
红线连接相邻的神经元。
包含红线的区域中的颜色指示神经元之间的距离。
颜色越深,表示距离越大。
颜色越浅,表示距离越小。
出现四组浅色线段,由一些颜色较暗的线段界定。这种组合方式表明网络已将数据聚类为四个组。这四组可以在前面的权重位置图窗中看到。
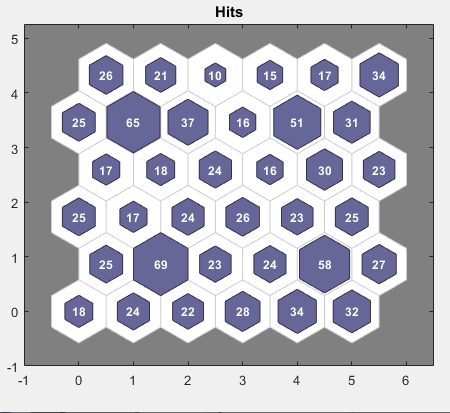
另一个有用的图窗可以显示每个神经元关联多少个数据点。点击 SOM 采样命中数可查看以下图窗。最佳情形是数据在神经元之间分布相当均匀。在此示例中,数据在边角处的神经元中稍微密集一些,但总体分布相当均匀。
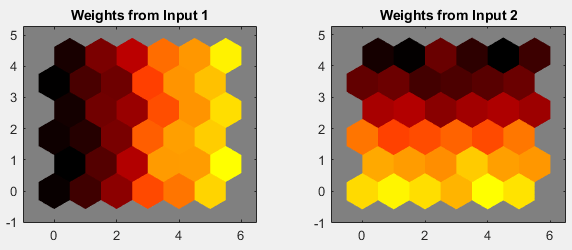
您也可以使用权重平面图窗来可视化权重本身。在训练窗口中点击 SOM 输入平面以获得下一个图窗。输入向量的每个元素有一个对应的权重平面(在本例中为两个)。它们是将每个输入连接到每个神经元的权重的可视化表示。(较浅和较深的颜色分别表示较大和较小的权重。)如果两个输入的连接模式非常相似,则可以假设这两个输入高度相关。在这种情况下,输入 1 的连接与输入 2 的连接非常不同。
您还可以从命令行生成前面的所有图窗。尝试下列绘图命令:plotsomhits、plotsomnc、plotsomnd、plotsomplanes、plotsompos 和 plotsomtop。