Medical Image-Based Finite Element Analysis of Spine
This example shows how to estimate bone stress and strain in a vertebra bone under axial compression using finite element (FE) analysis.
Bones experience mechanical loading due to gravity, motion, and muscle contraction. Estimating the stresses and strains within bone tissue can help predict bone strength and fracture risk. This example uses image-based FE analysis to predict bone stress and strain within a vertebra under axial compression. Image-based FE uses medical images, such as computed tomography (CT) scans, to generate the geometry and material properties of the FE model.
In this example, you create and analyze a 3-D model of a single vertebra under axial loading using this workflow:
Download Data
This example uses a CT scan saved as a directory of DICOM files. The scan is part of a data set that contains three CT volumes. Run this code to download the data set from the MathWorks website as a zip file and unzip the file. The total size of the data set is approximately 81 MB.
zipFile = matlab.internal.examples.downloadSupportFile("medical","MedicalVolumeDICOMData.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
After you download and unzip the data set, the scan used in this example is available in the dataFolder directory.
dataFolder = fullfile(filepath,"MedicalVolumeDICOMData","LungCT01");
Segment Vertebra from CT Scan
This example uses a binary segmentation mask of one vertebra in the spine. The mask was created by segmenting the spine from a chest CT scan using the Medical Image Labeler app. For an example of how to segment medical image volumes in the app, see Label 3-D Medical Image Using Medical Image Labeler.

The segmentation mask is attached to this example as a supporting file. Load the mask as a medicalVolume object.
labelVol = medicalVolume("LungCT01.nii.gz");The VolumeGeometry property of a medical volume object contains a medicalref3d object that defines the patient coordinate system for the scan. Extract the medicalref3d object for the mask into a new variable, R.
R = labelVol.VolumeGeometry;
Extract Vertebra Geometry and Generate FE Mesh
Extract Isosurface of Vertebra Mask
Calculate the isosurface faces and vertices for the vertebra mask by using the extractIsosurface function. The vertices coordinates are in the intrinsic coordinate system defined by rows, columns, and slices.
isovalue = 1; [faces,vertices] = extractIsosurface(labelVol.Voxels,isovalue);
Transform vertices to the patient coordinate system defined by R by using the intrinsicToWorld object function. The X, Y, and Z outputs define the xyz-coordinates of the surface points, in millimeters.
I = vertices(:,1); J = vertices(:,2); K = vertices(:,3); [X,Y,Z] = intrinsicToWorld(R,I,J,K); verticesPatient = [X Y Z];
Convert the vertex coordinates to meters.
verticesPatientMeters = verticesPatient.*10^-3;
Display the vertebral isosurface as a triangulated Surface object.
triangul = triangulation(double(faces),double(verticesPatientMeters)); viewer = viewer3d; surface = images.ui.graphics.Surface(viewer,data=triangul,Color=[1 0 0],Alpha=1);

Create PDE Model Geometry
Create a general PDEModel model container for a system of three equations. A general model allows you to assign spatially varying material properties based on bone density.
model = createpde(3);
Specify the geometry for the model by using the geometryFromMesh (Partial Differential Equation Toolbox) object function, with the isosurface in patient coordinates as input.
geometryFromMesh(model,verticesPatientMeters',faces');
pdegplot(model,FaceLabels="on")
Generate Mesh from Model Geometry
Specify the maximum edge length for the FE mesh elements. This example uses a max edge length of 1.6 mm, which was determined using a mesh convergence analysis.
hMax = 0.0016;
Generate the mesh from the model geometry. Specify the maximum edge length, and specify that the minimum edge length is half of the maximum edge length. The elements are quadratic tetrahedra.
msh = generateMesh(model,Hmax=hMax,Hmin=hMax/2); pdemesh(model);

Assign Material Properties
Bone material properties vary based on spatial location and direction:
Spatial location — Local regions with greater bone density are stiffer than low density regions.
Direction — Bone stiffness depends on the loading direction. This example models bone using transverse isotropy. A transversely isotropic material is stiffer in the axial direction versus in the transverse plane, and is uniform within the transverse plane.
In this example, you assign spatially varying, transversely isotropic linear elastic material properties.
Extract HU Values Within Vertebra Mask
To assign spatially varying material properties, you need to map CT intensity values to FE mesh coordinates.
Load the original, unsegmented CT scan using a medicalVolume object. The medicalVolume object automatically converts the intensity to Hounsfield units (HU). The HU value is a standard for measuring radiodensity.
DCMData = medicalVolume(dataFolder);
Extract the indices and intensities of CT voxels inside the vertebra mask.
trueIdx = find(labelVol.Voxels==1); HUVertebra = double(DCMData.Voxels(trueIdx));
Convert the linear indices in trueIdx into subscript indices, in units of voxels.
[row,col,slice] = ind2sub(size(labelVol.Voxels),trueIdx);
Transform the subscript indices into patient coordinates and convert the coordinates from millimeters to meters.
[X2,Y2,Z2] = intrinsicToWorld(R,col,row,slice); HUVertebraMeters = [X2 Y2 Z2].*10^-3;
Map HU Values from CT Voxels to Mesh Nodes
The FE node coordinates are scattered and not aligned with the CT voxel grid. Create a scatteredInterpolant object to define the 3-D interpolation between the voxel grid and FE nodes.
F = scatteredInterpolant(HUVertebraMeters,HUVertebra);
Specify Coefficients
Specify the PDE coefficients for the model. In this structural analysis, the c coefficient defines the material stiffness of the model, which is inversely related to compliance. To define a spatially varying c coefficient in Partial Differential Equation Toolbox™, represent c in function form. In this example, the HU2TransverseIsotropy helper function defines transversely isotropic material properties based on bone density. Bone density is calculated for a given location using the scattered interpolant F. The helper function is wrapped in an anonymous function, ccoeffunc, which passes the location and state structures to HU2TransverseIsotropy. The FE solver automatically computes location and state structure arrays and passes them to your function during the simulation.
ccoeffunc = @(location,state) HU2TransverseIsotropy(location,state,F); specifyCoefficients(model,'m',0, ... 'd',0, ... 'c',ccoeffunc, ... 'a',0, ... 'f',[0;0;0]);
Apply Loading and Solve Model
To simulate axial loading of the vertebra, fix the bottom surface and apply a downward load to the top surface. To simulate distributed loading, the load is applied as a pressure.
Identify the faces in the model geometry to apply the boundary conditions. In this example, the faces were identified using visual inspection of the plot created above using pdegplot.
bottomSurfaceFace = 1; topSurfaceFace = 250;
Specify the total force to apply, in newtons.
forceInput = -3000;
Estimate the area of the top surface.
nf2=findNodes(model.Mesh,"region",Face=topSurfaceFace);
positions = model.Mesh.Nodes(:,nf2)';
surfaceShape = alphaShape(positions(:,1:2));
faceArea = area(surfaceShape);Calculate the pressure magnitude as a function of force and area, in pascals.
inputPressure_Pa = forceInput/faceArea;
Apply the boundary conditions.
applyBoundaryCondition(model,"dirichlet",Face=bottomSurfaceFace,u=[0,0,0]); applyBoundaryCondition(model,"neumann",Face=topSurfaceFace,g=[0;0;inputPressure_Pa]);
Solve the model. The output is a StationaryResults (Partial Differential Equation Toolbox) object that contains nodal displacements and their gradients.
Rs = solvepde(model);
Analyze Results
View the results of the simulation by plotting the axial displacement, stress, and strain within the model.
Displacement
Plot axial displacement, in millimeters, for the full model by using the pdeplot3D function.
Uz = Rs.NodalSolution(:,3)*10^3; pdeplot3D(model,"ColorMapData",Uz) clim([-0.15 0]) title("Axial Displacement (mm)")

Plot displacement in a transverse slice at the midpoint of vertebral height.
Create a rectangular grid that covers the geometry of the transverse (xy) slice with spacing of 1 mm in each direction. The x and y vectors define the spatial limits and spacing in the transverse plane, and z is a scalar the provides the midpoint of vertebral height. The Xg, Yg, and Zg variables define the grid coordinates.
x = min(msh.Nodes(1,:)):0.001:max(msh.Nodes(1,:)); y = min(msh.Nodes(2,:)):0.001:max(msh.Nodes(2,:)); z = min(msh.Nodes(3,:))+0.5*(max(msh.Nodes(3,:))-min(msh.Nodes(3,:))); [Xg,Yg] = meshgrid(x,y); Zg = z*ones(size(Xg));
Interpolate normal axial displacement onto the grid coordinates by using the interpolateSolution (Partial Differential Equation Toolbox) object function. Convert displacement from meters to millimeters, and reshape the displacement vector to the grid size.
U = interpolateSolution(Rs,Xg,Yg,Zg,3); U = U*10^3; Ug = reshape(U,size(Xg));
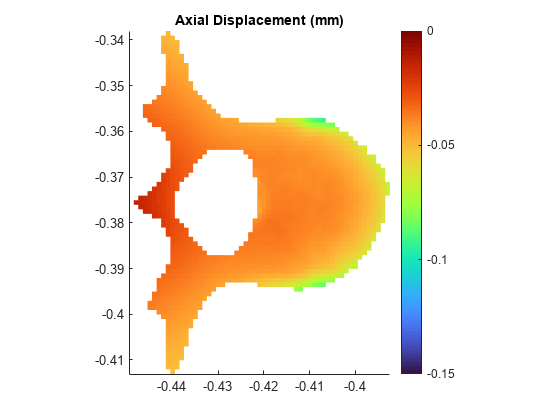
Plot axial displacement in the transverse slice.
surf(Xg,Yg,Ug,LineStyle="none") axis equal grid off view(0,90) colormap("turbo") colorbar clim([-0.15 0]) title("Axial Displacement (mm)")
Stress
In a structural analysis, stress is the product of the c coefficient and the gradients of displacement. Calculate normal stresses at the transverse slice grid coordinates by using the evaluateCGradient (Partial Differential Equation Toolbox) object function.
[cgradx,cgrady,cgradz] = evaluateCGradient(Rs,Xg,Yg,Zg,3);
Convert normal axial stress to megapascals, and reshape the stress vector to the grid size.
cgradz = cgradz*10^-6; cgradzg = reshape(cgradz,size(Xg));
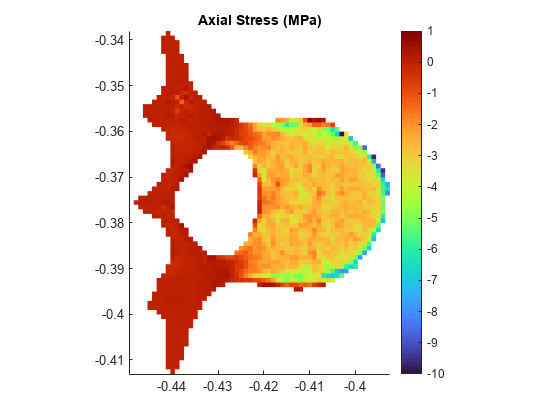
Plot the normal axial stress in the transverse slice.
surf(Xg,Yg,cgradzg,LineStyle="none"); axis equal grid off view(0,90) colormap("turbo") colorbar clim([-10 1]) title("Axial Stress (MPa)")
Strain
In a structural analysis, strain is the gradient of the displacement result. Extract the normal strain at the transverse slice grid coordinates by using the evaluateGradient (Partial Differential Equation Toolbox) object function.
[gradx,grady,gradz] = evaluateGradient(Rs,Xg,Yg,Zg,3);
Reshape the axial strain vector to the grid size.
gradzg = reshape(gradz,size(Xg));
Plot the axial strain in the transverse slice.
surf(Xg,Yg,gradzg,LineStyle="none"); axis equal grid off view(0,90) colormap("turbo") colorbar clim([-0.008 0]) title("Axial Strain")

Helper Functions
The HU2TransverseIsotropy helper function specifies transverse isotropic material properties using these steps:
Map voxel intensity to Hounsfield units (HU) at nodal locations using the
scatteredInterpolantobject,F.Convert HU to CT density using a linear calibration equation from [1].
Convert CT density to elastic modulus in the axial direction,
E3, and in the transverse plane,E12, as well as the bulk modulus,G, using density-elasticity relationships from [2]. The Poisson's ratio in the axial direction,v3, and transverse plane,v12, are also assigned based on [2].Define the 6-by-6 compliance matrix for transverse isotropy by using the
elasticityTransOrthoC3Dhelper function.Convert the 6-by-6 compliance, or c-matrix, to the vector form required by Partial Differential Equation Toolbox by using the
SixMat2NineMatandSquareMat2CCoeffVechelper functions.
function ccoeff = HU2TransverseIsotropy(location,state,F) HU = F(location.x,location.y,location.z); rho = 5.2+0.8*HU; E3 = -34.7 + 3.230.*rho; E12 = 0.333*E3; v12 = 0.104; v3 = 0.381; G = 0.121*E3; ccoeff = zeros(45,length(location.x)); for i = 1:length(location.x) cMatrix = elasticityTransOrthoC3D(E12(i),E3(i),v12,v3,G(i)); nineMat = SixMat2NineMat(cMatrix); ccoeff(:,i) = SquareMat2CCoeffVec(nineMat); end end
The elasticityTransOrthoC3D helper function defines the 6-by-6 compliance matrix for a transversely isotropic linear elastic material based on the elastic moduli, E12 and E3, bulk modulus, G, and Poisson's ratios, v12 and v3. The helper function converts all modulus values from megapascals to pascals before computing the compliance matrix.
function C = elasticityTransOrthoC3D(E12,E3,v12,v3,G) E12 = E12*10^6; E3 = E3*10^6; G = G*10^6; v_zp = (E3*v3)/E12; C = zeros(6,6); C(1,1) = 1/E12; C(1,2) = -v12/E12; C(1,3)= -v_zp/E3; C(2,1) = C(1,2); C(2,2) = 1/E12; C(2,3) = -v_zp/E3; C(3,1) = -v3/E12; C(3,2) = C(2,3); C(3,3) = 1/E3; C(4,4) = 1/(2*G); C(5,5) = 1/(2*G); C(6,6) = (1+v12)/E12; C=inv(C); end
The SixMat2NineMat helper function converts a 6-by-6 c coefficient matrix in Voigt notation to a 9-by-9 matrix corresponding to expanded form.
function nineMat = SixMat2NineMat(sixMat) for i = 1:6 nineVecs(i,:) = sixMat(i,[1 6 5 6 2 4 5 4 3]); end nineMat = [ nineVecs(1,:); ... nineVecs(6,:); ... nineVecs(5,:); ... nineVecs(6,:); ... nineVecs(2,:); ... nineVecs(4,:); ... nineVecs(5,:); ... nineVecs(4,:); ... nineVecs(3,:)]; end
The SquareMat2CCoeffVec converts a 9-by-9 c coefficient matrix into vector form as required by Partial Differential Equation Toolbox. This helper function creates a 45-element vector, corresponding to the "3N(3N+1)/2-Element Column Vector c, 3-D Systems" case in c Coefficient for specifyCoefficients (Partial Differential Equation Toolbox).
function c45Vec = SquareMat2CCoeffVec(nineMat) C11 = [nineMat(1,1) nineMat(1,2) nineMat(2,2) nineMat(1,3) nineMat(2,3) nineMat(3,3)]; C12 = [nineMat(1,4) nineMat(2,4) nineMat(3,4) nineMat(1,5) nineMat(2,5) nineMat(3,5) ... nineMat(1,6) nineMat(2,6) nineMat(3,6)]; C13 = [nineMat(1,7) nineMat(2,7) nineMat(3,7) nineMat(1,8) nineMat(2,8) nineMat(3,8) ... nineMat(1,9) nineMat(2,9) nineMat(3,9)]; C22 = [nineMat(4,4) nineMat(4,5) nineMat(5,5) nineMat(4,6) nineMat(5,6) nineMat(6,6)]; C23 = [nineMat(4,7) nineMat(5,7) nineMat(6,7) nineMat(4,8) nineMat(5,8) nineMat(6,8) ... nineMat(4,9) nineMat(5,9) nineMat(6,9)]; C33 = [nineMat(7,7) nineMat(7,8) nineMat(8,8) nineMat(7,9) nineMat(8,9) nineMat(9,9)]; c45Vec = [C11 C12 C22 C13 C23 C33]'; end
References
[1] Turbucz, Mate, Agoston Jakab Pokorni, György Szőke, Zoltan Hoffer, Rita Maria Kiss, Aron Lazary, and Peter Endre Eltes. “Development and Validation of Two Intact Lumbar Spine Finite Element Models for In Silico Investigations: Comparison of the Bone Modelling Approaches.” Applied Sciences 12, no. 20 (January 2022): 10256. https://doi.org/10.3390/app122010256.
[2] Unnikrishnan, Ginu U., and Elise F. Morgan. “A New Material Mapping Procedure for Quantitative Computed Tomography-Based, Continuum Finite Element Analyses of the Vertebra.” Journal of Biomechanical Engineering 133, no. 7 (July 1, 2011): 071001. https://doi.org/10.1115/1.4004190.
See Also
Medical Image
Labeler | medicalVolume | intrinsicToWorld | scatteredInterpolant | createpde (Partial Differential Equation Toolbox) | geometryFromMesh (Partial Differential Equation Toolbox) | solvepde (Partial Differential Equation Toolbox) | StationaryResults (Partial Differential Equation Toolbox)